Tutorial¶
This repository contains several utility functions that enable easier analysis across CMIP6 model data.
It offers solutions to the following problems:
Inconsistent naming of dimensions and coordinates
Inconsistent values,shape and dataset location of coordinates
Inconsistent longitude conventions
Inconsistent units
Inconsistent longitude/latitude bounds
TL;DR How to put it all together
[1]:
import matplotlib.pyplot as plt
import intake
import dask
%matplotlib inline
[2]:
url = "https://storage.googleapis.com/cmip6/pangeo-cmip6.json"
col = intake.open_esm_datastore(url)
## Inconsistent naming of dimensions and coordinates All cmip6 models (except for the unstructured grid of the AWI model) have in principal the same datastructure and should have a consistent naming, such that the user can test an analysis on one model and then seamlessly apply it on another. In practice some models have alternate naming for e.g. the logical (x,y,z) dimensions. xmip.preprocessing.rename_cmip6 accesses an internal dictionary to rename all models consistently to the
following scheme:
x,y,levfor the logical grid index in the x,y,z directionlon,latfor geographical position coordinatesbnds,vertexfor cell bounds or vertex indiciestime_bounds,lev_bounds,lon_bounds,lat_boundsfor cell bounding values
[3]:
# load a few models to illustrate the problem
query = dict(experiment_id=['piControl'], table_id='Oyr',
variable_id='o2', grid_label=['gn', 'gr'],
source_id=['IPSL-CM6A-LR', 'CanESM5', 'GFDL-ESM4']
)
cat = col.search(**query)
cat.df['source_id'].unique()
z_kwargs = {'consolidated': True, 'decode_times':False}
with dask.config.set(**{'array.slicing.split_large_chunks': True}):
dset_dict = cat.to_dataset_dict(zarr_kwargs=z_kwargs)#
--> The keys in the returned dictionary of datasets are constructed as follows:
'activity_id.institution_id.source_id.experiment_id.table_id.grid_label'
[4]:
# show coordinates
for k, ds in dset_dict.items():
print(k)
print(list(ds.dims))
CMIP.IPSL.IPSL-CM6A-LR.piControl.Oyr.gn
['axis_nbounds', 'member_id', 'nvertex', 'olevel', 'time', 'x', 'y']
CMIP.NOAA-GFDL.GFDL-ESM4.piControl.Oyr.gr
['bnds', 'lat', 'lev', 'lon', 'member_id', 'time']
CMIP.CCCma.CanESM5.piControl.Oyr.gn
['bnds', 'i', 'j', 'lev', 'member_id', 'time', 'vertices']
You can see that e.g. the x dimension is not consistently labelled. E.g. in one model it is called i in the other x. We can fix this by passing rename_cmip6 as preprocess argument to to_dataset_dict:
[5]:
from xmip.preprocessing import rename_cmip6
# load a few models to illustrate the problem
cat = col.search(**query)
cat.df['source_id'].unique()
# pass the preprocessing directly
with dask.config.set(**{'array.slicing.split_large_chunks': True}):
dset_dict_renamed = cat.to_dataset_dict(zarr_kwargs=z_kwargs, preprocess=rename_cmip6)
for k, ds in dset_dict_renamed.items():
print(k)
print(list(ds.dims))
--> The keys in the returned dictionary of datasets are constructed as follows:
'activity_id.institution_id.source_id.experiment_id.table_id.grid_label'
CMIP.IPSL.IPSL-CM6A-LR.piControl.Oyr.gn
['bnds', 'lev', 'member_id', 'time', 'vertex', 'x', 'y']
CMIP.NOAA-GFDL.GFDL-ESM4.piControl.Oyr.gr
['bnds', 'lev', 'member_id', 'time', 'x', 'y']
CMIP.CCCma.CanESM5.piControl.Oyr.gn
['bnds', 'lev', 'member_id', 'time', 'vertex', 'x', 'y']
Beautiful! They have exactly the same dimensions!

You can also always apply the utility functions after loading the data, but be aware that some models have even inconsistent namings between timesteps and ensemble members. This can cause problems with the concatenation that
intake_esmdoes. Passing the function will apply it before concatenation, which works nicely above. Here is an example of how it causes problems when applied afterwards:
[6]:
# IPSL data is a bit of a mess
ds = dset_dict['CMIP.IPSL.IPSL-CM6A-LR.piControl.Oyr.gn']
ds = rename_cmip6(ds)
ds
[6]:
<xarray.Dataset>
Dimensions: (bnds: 2, lev: 75, member_id: 1, time: 250, vertex: 4, x: 362, y: 332)
Coordinates:
lat_bounds (y, x, vertex) float32 dask.array<chunksize=(332, 362, 4), meta=np.ndarray>
lon_bounds (y, x, vertex) float32 dask.array<chunksize=(332, 362, 4), meta=np.ndarray>
lat (y, x) float32 dask.array<chunksize=(332, 362), meta=np.ndarray>
lon (y, x) float32 dask.array<chunksize=(332, 362), meta=np.ndarray>
* lev (lev) float32 0.5058 1.556 2.668 ... 5.698e+03 5.902e+03
lev_bounds (lev, bnds) float32 dask.array<chunksize=(75, 2), meta=np.ndarray>
* time (time) int64 0 8760 17532 26304 ... 2165184 2173944 2182704
time_bounds (time, bnds) float64 dask.array<chunksize=(250, 2), meta=np.ndarray>
* member_id (member_id) <U8 'r1i2p1f1'
Dimensions without coordinates: bnds, vertex, x, y
Data variables:
area (y, x) float32 dask.array<chunksize=(332, 362), meta=np.ndarray>
o2 (member_id, time, lev, y, x) float32 dask.array<chunksize=(1, 9, 75, 332, 362), meta=np.ndarray>
Attributes:
CMIP6_CV_version: cv=6.2.15.1
Conventions: CF-1.7 CMIP-6.2
EXPID: piControl
activity_id: CMIP
branch_method: standard
branch_time_in_child: 0.0
branch_time_in_parent: 36524.0
contact: ipsl-cmip6@listes.ipsl.fr
creation_date: 2019-02-11T12:01:20Z
data_specs_version: 01.00.28
description: DECK: control
dr2xml_md5sum: c2dce418e78ca835be1e2ff817c2c403
dr2xml_version: 1.16
experiment: pre-industrial control
experiment_id: piControl
external_variables: areacello volcello
forcing_index: 1
frequency: yr
further_info_url: https://furtherinfo.es-doc.org/CMIP6.IPSL.IPSL-C...
grid: native ocean tri-polar grid with 105 k ocean cells
grid_label: gn
history: none
initialization_index: 2
institution: Institut Pierre Simon Laplace, Paris 75252, France
institution_id: IPSL
license: CMIP6 model data produced by IPSL is licensed un...
mip_era: CMIP6
model_version: 6.1.8
name: /ccc/work/cont003/gencmip6/lebasn/IGCM_OUT/IPSLC...
nominal_resolution: 100 km
parent_activity_id: CMIP
parent_experiment_id: piControl-spinup
parent_mip_era: CMIP6
parent_source_id: IPSL-CM6A-LR
parent_time_units: days since 1750-01-01 00:00:00
parent_variant_label: r1i2p1f1
physics_index: 1
product: model-output
realization_index: 1
realm: ocnBgchem
source: IPSL-CM6A-LR (2017): atmos: LMDZ (NPv6, N96; 14...
source_id: IPSL-CM6A-LR
source_type: AOGCM BGC
sub_experiment: none
sub_experiment_id: none
table_id: Oyr
title: IPSL-CM6A-LR model output prepared for CMIP6 / C...
tracking_id: hdl:21.14100/5fb53c12-afe1-499d-9039-590b6e6b150...
variable_id: o2
variant_info: Equivalent to r1i1p1f1 but started on a differen...
variant_label: r1i2p1f1
status: 2019-10-25;created;by nhn2@columbia.edu
netcdf_tracking_ids: hdl:21.14100/5fb53c12-afe1-499d-9039-590b6e6b150...
version_id: v20190319
intake_esm_varname: ['o2']
intake_esm_dataset_key: CMIP.IPSL.IPSL-CM6A-LR.piControl.Oyr.gn- bnds: 2
- lev: 75
- member_id: 1
- time: 250
- vertex: 4
- x: 362
- y: 332
- lat_bounds(y, x, vertex)float32dask.array<chunksize=(332, 362, 4), meta=np.ndarray>
Array Chunk Bytes 1.92 MB 1.92 MB Shape (332, 362, 4) (332, 362, 4) Count 2 Tasks 1 Chunks Type float32 numpy.ndarray - lon_bounds(y, x, vertex)float32dask.array<chunksize=(332, 362, 4), meta=np.ndarray>
Array Chunk Bytes 1.92 MB 1.92 MB Shape (332, 362, 4) (332, 362, 4) Count 2 Tasks 1 Chunks Type float32 numpy.ndarray - lat(y, x)float32dask.array<chunksize=(332, 362), meta=np.ndarray>
- bounds :
- bounds_nav_lat
- long_name :
- Latitude
- standard_name :
- latitude
- units :
- degrees_north
Array Chunk Bytes 480.74 kB 480.74 kB Shape (332, 362) (332, 362) Count 2 Tasks 1 Chunks Type float32 numpy.ndarray - lon(y, x)float32dask.array<chunksize=(332, 362), meta=np.ndarray>
- bounds :
- bounds_nav_lon
- long_name :
- Longitude
- standard_name :
- longitude
- units :
- degrees_east
Array Chunk Bytes 480.74 kB 480.74 kB Shape (332, 362) (332, 362) Count 2 Tasks 1 Chunks Type float32 numpy.ndarray - lev(lev)float320.5058 1.556 ... 5.902e+03
- bounds :
- olevel_bounds
- long_name :
- Vertical T levels
- name :
- olevel
- positive :
- down
- units :
- m
array([5.057600e-01, 1.555855e+00, 2.667682e+00, 3.856280e+00, 5.140361e+00, 6.543034e+00, 8.092519e+00, 9.822750e+00, 1.177368e+01, 1.399104e+01, 1.652532e+01, 1.942980e+01, 2.275762e+01, 2.655830e+01, 3.087456e+01, 3.574020e+01, 4.118002e+01, 4.721189e+01, 5.385064e+01, 6.111284e+01, 6.902168e+01, 7.761116e+01, 8.692943e+01, 9.704131e+01, 1.080303e+02, 1.200000e+02, 1.330758e+02, 1.474062e+02, 1.631645e+02, 1.805499e+02, 1.997900e+02, 2.211412e+02, 2.448906e+02, 2.713564e+02, 3.008875e+02, 3.338628e+02, 3.706885e+02, 4.117939e+02, 4.576256e+02, 5.086399e+02, 5.652923e+02, 6.280260e+02, 6.972587e+02, 7.733683e+02, 8.566790e+02, 9.474479e+02, 1.045854e+03, 1.151991e+03, 1.265861e+03, 1.387377e+03, 1.516364e+03, 1.652568e+03, 1.795671e+03, 1.945296e+03, 2.101027e+03, 2.262422e+03, 2.429025e+03, 2.600380e+03, 2.776039e+03, 2.955570e+03, 3.138565e+03, 3.324641e+03, 3.513446e+03, 3.704657e+03, 3.897982e+03, 4.093159e+03, 4.289953e+03, 4.488155e+03, 4.687581e+03, 4.888070e+03, 5.089479e+03, 5.291683e+03, 5.494575e+03, 5.698061e+03, 5.902058e+03], dtype=float32) - lev_bounds(lev, bnds)float32dask.array<chunksize=(75, 2), meta=np.ndarray>
- units :
- m
Array Chunk Bytes 600 B 600 B Shape (75, 2) (75, 2) Count 2 Tasks 1 Chunks Type float32 numpy.ndarray - time(time)int640 8760 17532 ... 2173944 2182704
- axis :
- T
- bounds :
- time_bounds
- calendar :
- gregorian
- long_name :
- Time axis
- standard_name :
- time
- time_origin :
- 1850-01-01 00:00:00
- units :
- hours since 1850-07-02 12:00:00.000000
array([ 0, 8760, 17532, ..., 2165184, 2173944, 2182704])
- time_bounds(time, bnds)float64dask.array<chunksize=(250, 2), meta=np.ndarray>
- calendar :
- gregorian
- units :
- days since 1850-01-01
Array Chunk Bytes 4.00 kB 4.00 kB Shape (250, 2) (250, 2) Count 2 Tasks 1 Chunks Type float64 numpy.ndarray - member_id(member_id)<U8'r1i2p1f1'
array(['r1i2p1f1'], dtype='<U8')
- area(y, x)float32dask.array<chunksize=(332, 362), meta=np.ndarray>
- standard_name :
- cell_area
- units :
- m2
Array Chunk Bytes 480.74 kB 480.74 kB Shape (332, 362) (332, 362) Count 2 Tasks 1 Chunks Type float32 numpy.ndarray - o2(member_id, time, lev, y, x)float32dask.array<chunksize=(1, 9, 75, 332, 362), meta=np.ndarray>
- cell_measures :
- area: areacello volume: volcello
- cell_methods :
- area: mean where sea time: mean
- description :
- 'Mole concentration' means number of moles per unit volume, also called"molarity", and is used in the construction mole_concentration_of_X_in_Y, whereX is a material constituent of Y. A chemical or biological species denoted by X may be described by a single term such as 'nitrogen' or a phrase such as 'nox_expressed_as_nitrogen'.
- history :
- none
- interval_operation :
- 2700 s
- interval_write :
- 1 yr
- long_name :
- Dissolved Oxygen Concentration
- online_operation :
- average
- standard_name :
- mole_concentration_of_dissolved_molecular_oxygen_in_sea_water
- units :
- mol m-3
Array Chunk Bytes 9.01 GB 324.50 MB Shape (1, 250, 75, 332, 362) (1, 9, 75, 332, 362) Count 57 Tasks 28 Chunks Type float32 numpy.ndarray
- CMIP6_CV_version :
- cv=6.2.15.1
- Conventions :
- CF-1.7 CMIP-6.2
- EXPID :
- piControl
- activity_id :
- CMIP
- branch_method :
- standard
- branch_time_in_child :
- 0.0
- branch_time_in_parent :
- 36524.0
- contact :
- ipsl-cmip6@listes.ipsl.fr
- creation_date :
- 2019-02-11T12:01:20Z
- data_specs_version :
- 01.00.28
- description :
- DECK: control
- dr2xml_md5sum :
- c2dce418e78ca835be1e2ff817c2c403
- dr2xml_version :
- 1.16
- experiment :
- pre-industrial control
- experiment_id :
- piControl
- external_variables :
- areacello volcello
- forcing_index :
- 1
- frequency :
- yr
- further_info_url :
- https://furtherinfo.es-doc.org/CMIP6.IPSL.IPSL-CM6A-LR.piControl.none.r1i2p1f1
- grid :
- native ocean tri-polar grid with 105 k ocean cells
- grid_label :
- gn
- history :
- none
- initialization_index :
- 2
- institution :
- Institut Pierre Simon Laplace, Paris 75252, France
- institution_id :
- IPSL
- license :
- CMIP6 model data produced by IPSL is licensed under a Creative Commons Attribution-NonCommercial-ShareAlike 4.0 International License (https://creativecommons.org/licenses). Consult https://pcmdi.llnl.gov/CMIP6/TermsOfUse for terms of use governing CMIP6 output, including citation requirements and proper acknowledgment. Further information about this data, including some limitations, can be found via the further_info_url (recorded as a global attribute in this file) and at https://cmc.ipsl.fr/. The data producers and data providers make no warranty, either express or implied, including, but not limited to, warranties of merchantability and fitness for a particular purpose. All liabilities arising from the supply of the information (including any liability arising in negligence) are excluded to the fullest extent permitted by law.
- mip_era :
- CMIP6
- model_version :
- 6.1.8
- name :
- /ccc/work/cont003/gencmip6/lebasn/IGCM_OUT/IPSLCM6/PROD/piControl/CM61-LR-pi-i2/CMIP6/OCE/o2_Oyr_IPSL-CM6A-LR_piControl_r1i2p1f1_gn_%start_date%-%end_date%
- nominal_resolution :
- 100 km
- parent_activity_id :
- CMIP
- parent_experiment_id :
- piControl-spinup
- parent_mip_era :
- CMIP6
- parent_source_id :
- IPSL-CM6A-LR
- parent_time_units :
- days since 1750-01-01 00:00:00
- parent_variant_label :
- r1i2p1f1
- physics_index :
- 1
- product :
- model-output
- realization_index :
- 1
- realm :
- ocnBgchem
- source :
- IPSL-CM6A-LR (2017): atmos: LMDZ (NPv6, N96; 144 x 143 longitude/latitude; 79 levels; top level 40000 m) land: ORCHIDEE (v2.0, Water/Carbon/Energy mode) ocean: NEMO-OPA (eORCA1.3, tripolar primarily 1deg; 362 x 332 longitude/latitude; 75 levels; top grid cell 0-2 m) ocnBgchem: NEMO-PISCES seaIce: NEMO-LIM3
- source_id :
- IPSL-CM6A-LR
- source_type :
- AOGCM BGC
- sub_experiment :
- none
- sub_experiment_id :
- none
- table_id :
- Oyr
- title :
- IPSL-CM6A-LR model output prepared for CMIP6 / CMIP piControl
- tracking_id :
- hdl:21.14100/5fb53c12-afe1-499d-9039-590b6e6b1501 hdl:21.14100/0825a99b-e57e-4796-9f3b-0d260c960c30
- variable_id :
- o2
- variant_info :
- Equivalent to r1i1p1f1 but started on a different machine. Information provided by this attribute may in some cases be flawed. Users can find more comprehensive and up-to-date documentation via the further_info_url global attribute.
- variant_label :
- r1i2p1f1
- status :
- 2019-10-25;created;by nhn2@columbia.edu
- netcdf_tracking_ids :
- hdl:21.14100/5fb53c12-afe1-499d-9039-590b6e6b1501 hdl:21.14100/0825a99b-e57e-4796-9f3b-0d260c960c30
- version_id :
- v20190319
- intake_esm_varname :
- ['o2']
- intake_esm_dataset_key :
- CMIP.IPSL.IPSL-CM6A-LR.piControl.Oyr.gn
~See how the data_variable o2 has several depth variables o2(member_id, time, olevel, y, x, lev)~ > This has recently been fixed in the pangeo google store, but does still apply if you are e.g. working with a local copy of the CMIP6 netdcf file.
I strongly recommend applying all preprocessing using the ``preprocess`` keyword, but it is crucial to do so with the initial renaming step
## Inconsistent values,shape and dataset location of coordinates The naming of the dimensions/coordinates is only the beginning: Some datasets use only index values for the x,y dimensions, while others use nominal longitudes, latitudes (usefull for rough region selection) or the longitudes and latitudes are only 1d arrays (e.g. for regridded outputs). Our goal is to work with all datasets int the same way, and hence we convert all datasets in this form:
x,yarea given as 1D ‘nominal’ longitudes and latitudes. This means thexis the zonal average latitude (can become difficult near the Arctic, but is otherwise very useful) andyis the longitude at the equator.lonandlatare 2-dimensional coordinate arrays with the ‘true’ position of grid cells (if the values were initially given as 1d arrays, they are broadcasted appropriately)
We achieve this by applying promote_empty_dims (give empty dimensions values), broadcast_lonlat (convert 1d lon and lat arrays to 2d arrays) and replace_x_y_nominal_lat_lon (calculate nominal lon and lat and replace x and y with them)
[7]:
from xmip.preprocessing import promote_empty_dims, broadcast_lonlat, replace_x_y_nominal_lat_lon
# check out the previous datasets
ds = dset_dict_renamed['CMIP.IPSL.IPSL-CM6A-LR.piControl.Oyr.gn']
ds
[7]:
<xarray.Dataset>
Dimensions: (bnds: 2, lev: 75, member_id: 1, time: 250, vertex: 4, x: 362, y: 332)
Coordinates:
lat_bounds (y, x, vertex) float32 dask.array<chunksize=(332, 362, 4), meta=np.ndarray>
lon_bounds (y, x, vertex) float32 dask.array<chunksize=(332, 362, 4), meta=np.ndarray>
lat (y, x) float32 dask.array<chunksize=(332, 362), meta=np.ndarray>
lon (y, x) float32 dask.array<chunksize=(332, 362), meta=np.ndarray>
* lev (lev) float32 0.5058 1.556 2.668 ... 5.698e+03 5.902e+03
lev_bounds (lev, bnds) float32 dask.array<chunksize=(75, 2), meta=np.ndarray>
* time (time) int64 0 8760 17532 26304 ... 2165184 2173944 2182704
time_bounds (time, bnds) float64 dask.array<chunksize=(250, 2), meta=np.ndarray>
* member_id (member_id) <U8 'r1i2p1f1'
Dimensions without coordinates: bnds, vertex, x, y
Data variables:
area (y, x) float32 dask.array<chunksize=(332, 362), meta=np.ndarray>
o2 (member_id, time, lev, y, x) float32 dask.array<chunksize=(1, 9, 75, 332, 362), meta=np.ndarray>
Attributes:
CMIP6_CV_version: cv=6.2.15.1
Conventions: CF-1.7 CMIP-6.2
EXPID: piControl
activity_id: CMIP
branch_method: standard
branch_time_in_child: 0.0
branch_time_in_parent: 36524.0
contact: ipsl-cmip6@listes.ipsl.fr
creation_date: 2019-02-11T12:01:20Z
data_specs_version: 01.00.28
description: DECK: control
dr2xml_md5sum: c2dce418e78ca835be1e2ff817c2c403
dr2xml_version: 1.16
experiment: pre-industrial control
experiment_id: piControl
external_variables: areacello volcello
forcing_index: 1
frequency: yr
further_info_url: https://furtherinfo.es-doc.org/CMIP6.IPSL.IPSL-C...
grid: native ocean tri-polar grid with 105 k ocean cells
grid_label: gn
history: none
initialization_index: 2
institution: Institut Pierre Simon Laplace, Paris 75252, France
institution_id: IPSL
license: CMIP6 model data produced by IPSL is licensed un...
mip_era: CMIP6
model_version: 6.1.8
name: /ccc/work/cont003/gencmip6/lebasn/IGCM_OUT/IPSLC...
nominal_resolution: 100 km
parent_activity_id: CMIP
parent_experiment_id: piControl-spinup
parent_mip_era: CMIP6
parent_source_id: IPSL-CM6A-LR
parent_time_units: days since 1750-01-01 00:00:00
parent_variant_label: r1i2p1f1
physics_index: 1
product: model-output
realization_index: 1
realm: ocnBgchem
source: IPSL-CM6A-LR (2017): atmos: LMDZ (NPv6, N96; 14...
source_id: IPSL-CM6A-LR
source_type: AOGCM BGC
sub_experiment: none
sub_experiment_id: none
table_id: Oyr
title: IPSL-CM6A-LR model output prepared for CMIP6 / C...
tracking_id: hdl:21.14100/5fb53c12-afe1-499d-9039-590b6e6b150...
variable_id: o2
variant_info: Equivalent to r1i1p1f1 but started on a differen...
variant_label: r1i2p1f1
status: 2019-10-25;created;by nhn2@columbia.edu
netcdf_tracking_ids: hdl:21.14100/5fb53c12-afe1-499d-9039-590b6e6b150...
version_id: v20190319
intake_esm_varname: ['o2']
intake_esm_dataset_key: CMIP.IPSL.IPSL-CM6A-LR.piControl.Oyr.gn- bnds: 2
- lev: 75
- member_id: 1
- time: 250
- vertex: 4
- x: 362
- y: 332
- lat_bounds(y, x, vertex)float32dask.array<chunksize=(332, 362, 4), meta=np.ndarray>
Array Chunk Bytes 1.92 MB 1.92 MB Shape (332, 362, 4) (332, 362, 4) Count 2 Tasks 1 Chunks Type float32 numpy.ndarray - lon_bounds(y, x, vertex)float32dask.array<chunksize=(332, 362, 4), meta=np.ndarray>
Array Chunk Bytes 1.92 MB 1.92 MB Shape (332, 362, 4) (332, 362, 4) Count 2 Tasks 1 Chunks Type float32 numpy.ndarray - lat(y, x)float32dask.array<chunksize=(332, 362), meta=np.ndarray>
- bounds :
- bounds_nav_lat
- long_name :
- Latitude
- standard_name :
- latitude
- units :
- degrees_north
Array Chunk Bytes 480.74 kB 480.74 kB Shape (332, 362) (332, 362) Count 2 Tasks 1 Chunks Type float32 numpy.ndarray - lon(y, x)float32dask.array<chunksize=(332, 362), meta=np.ndarray>
- bounds :
- bounds_nav_lon
- long_name :
- Longitude
- standard_name :
- longitude
- units :
- degrees_east
Array Chunk Bytes 480.74 kB 480.74 kB Shape (332, 362) (332, 362) Count 2 Tasks 1 Chunks Type float32 numpy.ndarray - lev(lev)float320.5058 1.556 ... 5.902e+03
- bounds :
- olevel_bounds
- long_name :
- Vertical T levels
- name :
- olevel
- positive :
- down
- units :
- m
array([5.057600e-01, 1.555855e+00, 2.667682e+00, 3.856280e+00, 5.140361e+00, 6.543034e+00, 8.092519e+00, 9.822750e+00, 1.177368e+01, 1.399104e+01, 1.652532e+01, 1.942980e+01, 2.275762e+01, 2.655830e+01, 3.087456e+01, 3.574020e+01, 4.118002e+01, 4.721189e+01, 5.385064e+01, 6.111284e+01, 6.902168e+01, 7.761116e+01, 8.692943e+01, 9.704131e+01, 1.080303e+02, 1.200000e+02, 1.330758e+02, 1.474062e+02, 1.631645e+02, 1.805499e+02, 1.997900e+02, 2.211412e+02, 2.448906e+02, 2.713564e+02, 3.008875e+02, 3.338628e+02, 3.706885e+02, 4.117939e+02, 4.576256e+02, 5.086399e+02, 5.652923e+02, 6.280260e+02, 6.972587e+02, 7.733683e+02, 8.566790e+02, 9.474479e+02, 1.045854e+03, 1.151991e+03, 1.265861e+03, 1.387377e+03, 1.516364e+03, 1.652568e+03, 1.795671e+03, 1.945296e+03, 2.101027e+03, 2.262422e+03, 2.429025e+03, 2.600380e+03, 2.776039e+03, 2.955570e+03, 3.138565e+03, 3.324641e+03, 3.513446e+03, 3.704657e+03, 3.897982e+03, 4.093159e+03, 4.289953e+03, 4.488155e+03, 4.687581e+03, 4.888070e+03, 5.089479e+03, 5.291683e+03, 5.494575e+03, 5.698061e+03, 5.902058e+03], dtype=float32) - lev_bounds(lev, bnds)float32dask.array<chunksize=(75, 2), meta=np.ndarray>
- units :
- m
Array Chunk Bytes 600 B 600 B Shape (75, 2) (75, 2) Count 2 Tasks 1 Chunks Type float32 numpy.ndarray - time(time)int640 8760 17532 ... 2173944 2182704
- axis :
- T
- bounds :
- time_bounds
- calendar :
- gregorian
- long_name :
- Time axis
- standard_name :
- time
- time_origin :
- 1850-01-01 00:00:00
- units :
- hours since 1850-07-02 12:00:00.000000
array([ 0, 8760, 17532, ..., 2165184, 2173944, 2182704])
- time_bounds(time, bnds)float64dask.array<chunksize=(250, 2), meta=np.ndarray>
- calendar :
- gregorian
- units :
- days since 1850-01-01
Array Chunk Bytes 4.00 kB 4.00 kB Shape (250, 2) (250, 2) Count 2 Tasks 1 Chunks Type float64 numpy.ndarray - member_id(member_id)<U8'r1i2p1f1'
array(['r1i2p1f1'], dtype='<U8')
- area(y, x)float32dask.array<chunksize=(332, 362), meta=np.ndarray>
- standard_name :
- cell_area
- units :
- m2
Array Chunk Bytes 480.74 kB 480.74 kB Shape (332, 362) (332, 362) Count 2 Tasks 1 Chunks Type float32 numpy.ndarray - o2(member_id, time, lev, y, x)float32dask.array<chunksize=(1, 9, 75, 332, 362), meta=np.ndarray>
- cell_measures :
- area: areacello volume: volcello
- cell_methods :
- area: mean where sea time: mean
- description :
- 'Mole concentration' means number of moles per unit volume, also called"molarity", and is used in the construction mole_concentration_of_X_in_Y, whereX is a material constituent of Y. A chemical or biological species denoted by X may be described by a single term such as 'nitrogen' or a phrase such as 'nox_expressed_as_nitrogen'.
- history :
- none
- interval_operation :
- 2700 s
- interval_write :
- 1 yr
- long_name :
- Dissolved Oxygen Concentration
- online_operation :
- average
- standard_name :
- mole_concentration_of_dissolved_molecular_oxygen_in_sea_water
- units :
- mol m-3
Array Chunk Bytes 9.01 GB 324.50 MB Shape (1, 250, 75, 332, 362) (1, 9, 75, 332, 362) Count 57 Tasks 28 Chunks Type float32 numpy.ndarray
- CMIP6_CV_version :
- cv=6.2.15.1
- Conventions :
- CF-1.7 CMIP-6.2
- EXPID :
- piControl
- activity_id :
- CMIP
- branch_method :
- standard
- branch_time_in_child :
- 0.0
- branch_time_in_parent :
- 36524.0
- contact :
- ipsl-cmip6@listes.ipsl.fr
- creation_date :
- 2019-02-11T12:01:20Z
- data_specs_version :
- 01.00.28
- description :
- DECK: control
- dr2xml_md5sum :
- c2dce418e78ca835be1e2ff817c2c403
- dr2xml_version :
- 1.16
- experiment :
- pre-industrial control
- experiment_id :
- piControl
- external_variables :
- areacello volcello
- forcing_index :
- 1
- frequency :
- yr
- further_info_url :
- https://furtherinfo.es-doc.org/CMIP6.IPSL.IPSL-CM6A-LR.piControl.none.r1i2p1f1
- grid :
- native ocean tri-polar grid with 105 k ocean cells
- grid_label :
- gn
- history :
- none
- initialization_index :
- 2
- institution :
- Institut Pierre Simon Laplace, Paris 75252, France
- institution_id :
- IPSL
- license :
- CMIP6 model data produced by IPSL is licensed under a Creative Commons Attribution-NonCommercial-ShareAlike 4.0 International License (https://creativecommons.org/licenses). Consult https://pcmdi.llnl.gov/CMIP6/TermsOfUse for terms of use governing CMIP6 output, including citation requirements and proper acknowledgment. Further information about this data, including some limitations, can be found via the further_info_url (recorded as a global attribute in this file) and at https://cmc.ipsl.fr/. The data producers and data providers make no warranty, either express or implied, including, but not limited to, warranties of merchantability and fitness for a particular purpose. All liabilities arising from the supply of the information (including any liability arising in negligence) are excluded to the fullest extent permitted by law.
- mip_era :
- CMIP6
- model_version :
- 6.1.8
- name :
- /ccc/work/cont003/gencmip6/lebasn/IGCM_OUT/IPSLCM6/PROD/piControl/CM61-LR-pi-i2/CMIP6/OCE/o2_Oyr_IPSL-CM6A-LR_piControl_r1i2p1f1_gn_%start_date%-%end_date%
- nominal_resolution :
- 100 km
- parent_activity_id :
- CMIP
- parent_experiment_id :
- piControl-spinup
- parent_mip_era :
- CMIP6
- parent_source_id :
- IPSL-CM6A-LR
- parent_time_units :
- days since 1750-01-01 00:00:00
- parent_variant_label :
- r1i2p1f1
- physics_index :
- 1
- product :
- model-output
- realization_index :
- 1
- realm :
- ocnBgchem
- source :
- IPSL-CM6A-LR (2017): atmos: LMDZ (NPv6, N96; 144 x 143 longitude/latitude; 79 levels; top level 40000 m) land: ORCHIDEE (v2.0, Water/Carbon/Energy mode) ocean: NEMO-OPA (eORCA1.3, tripolar primarily 1deg; 362 x 332 longitude/latitude; 75 levels; top grid cell 0-2 m) ocnBgchem: NEMO-PISCES seaIce: NEMO-LIM3
- source_id :
- IPSL-CM6A-LR
- source_type :
- AOGCM BGC
- sub_experiment :
- none
- sub_experiment_id :
- none
- table_id :
- Oyr
- title :
- IPSL-CM6A-LR model output prepared for CMIP6 / CMIP piControl
- tracking_id :
- hdl:21.14100/5fb53c12-afe1-499d-9039-590b6e6b1501 hdl:21.14100/0825a99b-e57e-4796-9f3b-0d260c960c30
- variable_id :
- o2
- variant_info :
- Equivalent to r1i1p1f1 but started on a different machine. Information provided by this attribute may in some cases be flawed. Users can find more comprehensive and up-to-date documentation via the further_info_url global attribute.
- variant_label :
- r1i2p1f1
- status :
- 2019-10-25;created;by nhn2@columbia.edu
- netcdf_tracking_ids :
- hdl:21.14100/5fb53c12-afe1-499d-9039-590b6e6b1501 hdl:21.14100/0825a99b-e57e-4796-9f3b-0d260c960c30
- version_id :
- v20190319
- intake_esm_varname :
- ['o2']
- intake_esm_dataset_key :
- CMIP.IPSL.IPSL-CM6A-LR.piControl.Oyr.gn
Note how the dimensions x and y dont have values (e.g. are not listed as coords)
[8]:
ds = promote_empty_dims(ds)
ds
[8]:
<xarray.Dataset>
Dimensions: (bnds: 2, lev: 75, member_id: 1, time: 250, vertex: 4, x: 362, y: 332)
Coordinates:
lat_bounds (y, x, vertex) float32 dask.array<chunksize=(332, 362, 4), meta=np.ndarray>
lon_bounds (y, x, vertex) float32 dask.array<chunksize=(332, 362, 4), meta=np.ndarray>
lat (y, x) float32 dask.array<chunksize=(332, 362), meta=np.ndarray>
lon (y, x) float32 dask.array<chunksize=(332, 362), meta=np.ndarray>
* lev (lev) float32 0.5058 1.556 2.668 ... 5.698e+03 5.902e+03
lev_bounds (lev, bnds) float32 dask.array<chunksize=(75, 2), meta=np.ndarray>
* time (time) int64 0 8760 17532 26304 ... 2165184 2173944 2182704
time_bounds (time, bnds) float64 dask.array<chunksize=(250, 2), meta=np.ndarray>
* member_id (member_id) <U8 'r1i2p1f1'
* bnds (bnds) int64 0 1
* vertex (vertex) int64 0 1 2 3
* x (x) int64 0 1 2 3 4 5 6 7 8 ... 354 355 356 357 358 359 360 361
* y (y) int64 0 1 2 3 4 5 6 7 8 ... 324 325 326 327 328 329 330 331
Data variables:
area (y, x) float32 dask.array<chunksize=(332, 362), meta=np.ndarray>
o2 (member_id, time, lev, y, x) float32 dask.array<chunksize=(1, 9, 75, 332, 362), meta=np.ndarray>
Attributes:
CMIP6_CV_version: cv=6.2.15.1
Conventions: CF-1.7 CMIP-6.2
EXPID: piControl
activity_id: CMIP
branch_method: standard
branch_time_in_child: 0.0
branch_time_in_parent: 36524.0
contact: ipsl-cmip6@listes.ipsl.fr
creation_date: 2019-02-11T12:01:20Z
data_specs_version: 01.00.28
description: DECK: control
dr2xml_md5sum: c2dce418e78ca835be1e2ff817c2c403
dr2xml_version: 1.16
experiment: pre-industrial control
experiment_id: piControl
external_variables: areacello volcello
forcing_index: 1
frequency: yr
further_info_url: https://furtherinfo.es-doc.org/CMIP6.IPSL.IPSL-C...
grid: native ocean tri-polar grid with 105 k ocean cells
grid_label: gn
history: none
initialization_index: 2
institution: Institut Pierre Simon Laplace, Paris 75252, France
institution_id: IPSL
license: CMIP6 model data produced by IPSL is licensed un...
mip_era: CMIP6
model_version: 6.1.8
name: /ccc/work/cont003/gencmip6/lebasn/IGCM_OUT/IPSLC...
nominal_resolution: 100 km
parent_activity_id: CMIP
parent_experiment_id: piControl-spinup
parent_mip_era: CMIP6
parent_source_id: IPSL-CM6A-LR
parent_time_units: days since 1750-01-01 00:00:00
parent_variant_label: r1i2p1f1
physics_index: 1
product: model-output
realization_index: 1
realm: ocnBgchem
source: IPSL-CM6A-LR (2017): atmos: LMDZ (NPv6, N96; 14...
source_id: IPSL-CM6A-LR
source_type: AOGCM BGC
sub_experiment: none
sub_experiment_id: none
table_id: Oyr
title: IPSL-CM6A-LR model output prepared for CMIP6 / C...
tracking_id: hdl:21.14100/5fb53c12-afe1-499d-9039-590b6e6b150...
variable_id: o2
variant_info: Equivalent to r1i1p1f1 but started on a differen...
variant_label: r1i2p1f1
status: 2019-10-25;created;by nhn2@columbia.edu
netcdf_tracking_ids: hdl:21.14100/5fb53c12-afe1-499d-9039-590b6e6b150...
version_id: v20190319
intake_esm_varname: ['o2']
intake_esm_dataset_key: CMIP.IPSL.IPSL-CM6A-LR.piControl.Oyr.gn- bnds: 2
- lev: 75
- member_id: 1
- time: 250
- vertex: 4
- x: 362
- y: 332
- lat_bounds(y, x, vertex)float32dask.array<chunksize=(332, 362, 4), meta=np.ndarray>
Array Chunk Bytes 1.92 MB 1.92 MB Shape (332, 362, 4) (332, 362, 4) Count 2 Tasks 1 Chunks Type float32 numpy.ndarray - lon_bounds(y, x, vertex)float32dask.array<chunksize=(332, 362, 4), meta=np.ndarray>
Array Chunk Bytes 1.92 MB 1.92 MB Shape (332, 362, 4) (332, 362, 4) Count 2 Tasks 1 Chunks Type float32 numpy.ndarray - lat(y, x)float32dask.array<chunksize=(332, 362), meta=np.ndarray>
- bounds :
- bounds_nav_lat
- long_name :
- Latitude
- standard_name :
- latitude
- units :
- degrees_north
Array Chunk Bytes 480.74 kB 480.74 kB Shape (332, 362) (332, 362) Count 2 Tasks 1 Chunks Type float32 numpy.ndarray - lon(y, x)float32dask.array<chunksize=(332, 362), meta=np.ndarray>
- bounds :
- bounds_nav_lon
- long_name :
- Longitude
- standard_name :
- longitude
- units :
- degrees_east
Array Chunk Bytes 480.74 kB 480.74 kB Shape (332, 362) (332, 362) Count 2 Tasks 1 Chunks Type float32 numpy.ndarray - lev(lev)float320.5058 1.556 ... 5.902e+03
- bounds :
- olevel_bounds
- long_name :
- Vertical T levels
- name :
- olevel
- positive :
- down
- units :
- m
array([5.057600e-01, 1.555855e+00, 2.667682e+00, 3.856280e+00, 5.140361e+00, 6.543034e+00, 8.092519e+00, 9.822750e+00, 1.177368e+01, 1.399104e+01, 1.652532e+01, 1.942980e+01, 2.275762e+01, 2.655830e+01, 3.087456e+01, 3.574020e+01, 4.118002e+01, 4.721189e+01, 5.385064e+01, 6.111284e+01, 6.902168e+01, 7.761116e+01, 8.692943e+01, 9.704131e+01, 1.080303e+02, 1.200000e+02, 1.330758e+02, 1.474062e+02, 1.631645e+02, 1.805499e+02, 1.997900e+02, 2.211412e+02, 2.448906e+02, 2.713564e+02, 3.008875e+02, 3.338628e+02, 3.706885e+02, 4.117939e+02, 4.576256e+02, 5.086399e+02, 5.652923e+02, 6.280260e+02, 6.972587e+02, 7.733683e+02, 8.566790e+02, 9.474479e+02, 1.045854e+03, 1.151991e+03, 1.265861e+03, 1.387377e+03, 1.516364e+03, 1.652568e+03, 1.795671e+03, 1.945296e+03, 2.101027e+03, 2.262422e+03, 2.429025e+03, 2.600380e+03, 2.776039e+03, 2.955570e+03, 3.138565e+03, 3.324641e+03, 3.513446e+03, 3.704657e+03, 3.897982e+03, 4.093159e+03, 4.289953e+03, 4.488155e+03, 4.687581e+03, 4.888070e+03, 5.089479e+03, 5.291683e+03, 5.494575e+03, 5.698061e+03, 5.902058e+03], dtype=float32) - lev_bounds(lev, bnds)float32dask.array<chunksize=(75, 2), meta=np.ndarray>
- units :
- m
Array Chunk Bytes 600 B 600 B Shape (75, 2) (75, 2) Count 2 Tasks 1 Chunks Type float32 numpy.ndarray - time(time)int640 8760 17532 ... 2173944 2182704
- axis :
- T
- bounds :
- time_bounds
- calendar :
- gregorian
- long_name :
- Time axis
- standard_name :
- time
- time_origin :
- 1850-01-01 00:00:00
- units :
- hours since 1850-07-02 12:00:00.000000
array([ 0, 8760, 17532, ..., 2165184, 2173944, 2182704])
- time_bounds(time, bnds)float64dask.array<chunksize=(250, 2), meta=np.ndarray>
- calendar :
- gregorian
- units :
- days since 1850-01-01
Array Chunk Bytes 4.00 kB 4.00 kB Shape (250, 2) (250, 2) Count 2 Tasks 1 Chunks Type float64 numpy.ndarray - member_id(member_id)<U8'r1i2p1f1'
array(['r1i2p1f1'], dtype='<U8')
- bnds(bnds)int640 1
array([0, 1])
- vertex(vertex)int640 1 2 3
array([0, 1, 2, 3])
- x(x)int640 1 2 3 4 5 ... 357 358 359 360 361
array([ 0, 1, 2, ..., 359, 360, 361])
- y(y)int640 1 2 3 4 5 ... 327 328 329 330 331
array([ 0, 1, 2, ..., 329, 330, 331])
- area(y, x)float32dask.array<chunksize=(332, 362), meta=np.ndarray>
- standard_name :
- cell_area
- units :
- m2
Array Chunk Bytes 480.74 kB 480.74 kB Shape (332, 362) (332, 362) Count 2 Tasks 1 Chunks Type float32 numpy.ndarray - o2(member_id, time, lev, y, x)float32dask.array<chunksize=(1, 9, 75, 332, 362), meta=np.ndarray>
- cell_measures :
- area: areacello volume: volcello
- cell_methods :
- area: mean where sea time: mean
- description :
- 'Mole concentration' means number of moles per unit volume, also called"molarity", and is used in the construction mole_concentration_of_X_in_Y, whereX is a material constituent of Y. A chemical or biological species denoted by X may be described by a single term such as 'nitrogen' or a phrase such as 'nox_expressed_as_nitrogen'.
- history :
- none
- interval_operation :
- 2700 s
- interval_write :
- 1 yr
- long_name :
- Dissolved Oxygen Concentration
- online_operation :
- average
- standard_name :
- mole_concentration_of_dissolved_molecular_oxygen_in_sea_water
- units :
- mol m-3
Array Chunk Bytes 9.01 GB 324.50 MB Shape (1, 250, 75, 332, 362) (1, 9, 75, 332, 362) Count 57 Tasks 28 Chunks Type float32 numpy.ndarray
- CMIP6_CV_version :
- cv=6.2.15.1
- Conventions :
- CF-1.7 CMIP-6.2
- EXPID :
- piControl
- activity_id :
- CMIP
- branch_method :
- standard
- branch_time_in_child :
- 0.0
- branch_time_in_parent :
- 36524.0
- contact :
- ipsl-cmip6@listes.ipsl.fr
- creation_date :
- 2019-02-11T12:01:20Z
- data_specs_version :
- 01.00.28
- description :
- DECK: control
- dr2xml_md5sum :
- c2dce418e78ca835be1e2ff817c2c403
- dr2xml_version :
- 1.16
- experiment :
- pre-industrial control
- experiment_id :
- piControl
- external_variables :
- areacello volcello
- forcing_index :
- 1
- frequency :
- yr
- further_info_url :
- https://furtherinfo.es-doc.org/CMIP6.IPSL.IPSL-CM6A-LR.piControl.none.r1i2p1f1
- grid :
- native ocean tri-polar grid with 105 k ocean cells
- grid_label :
- gn
- history :
- none
- initialization_index :
- 2
- institution :
- Institut Pierre Simon Laplace, Paris 75252, France
- institution_id :
- IPSL
- license :
- CMIP6 model data produced by IPSL is licensed under a Creative Commons Attribution-NonCommercial-ShareAlike 4.0 International License (https://creativecommons.org/licenses). Consult https://pcmdi.llnl.gov/CMIP6/TermsOfUse for terms of use governing CMIP6 output, including citation requirements and proper acknowledgment. Further information about this data, including some limitations, can be found via the further_info_url (recorded as a global attribute in this file) and at https://cmc.ipsl.fr/. The data producers and data providers make no warranty, either express or implied, including, but not limited to, warranties of merchantability and fitness for a particular purpose. All liabilities arising from the supply of the information (including any liability arising in negligence) are excluded to the fullest extent permitted by law.
- mip_era :
- CMIP6
- model_version :
- 6.1.8
- name :
- /ccc/work/cont003/gencmip6/lebasn/IGCM_OUT/IPSLCM6/PROD/piControl/CM61-LR-pi-i2/CMIP6/OCE/o2_Oyr_IPSL-CM6A-LR_piControl_r1i2p1f1_gn_%start_date%-%end_date%
- nominal_resolution :
- 100 km
- parent_activity_id :
- CMIP
- parent_experiment_id :
- piControl-spinup
- parent_mip_era :
- CMIP6
- parent_source_id :
- IPSL-CM6A-LR
- parent_time_units :
- days since 1750-01-01 00:00:00
- parent_variant_label :
- r1i2p1f1
- physics_index :
- 1
- product :
- model-output
- realization_index :
- 1
- realm :
- ocnBgchem
- source :
- IPSL-CM6A-LR (2017): atmos: LMDZ (NPv6, N96; 144 x 143 longitude/latitude; 79 levels; top level 40000 m) land: ORCHIDEE (v2.0, Water/Carbon/Energy mode) ocean: NEMO-OPA (eORCA1.3, tripolar primarily 1deg; 362 x 332 longitude/latitude; 75 levels; top grid cell 0-2 m) ocnBgchem: NEMO-PISCES seaIce: NEMO-LIM3
- source_id :
- IPSL-CM6A-LR
- source_type :
- AOGCM BGC
- sub_experiment :
- none
- sub_experiment_id :
- none
- table_id :
- Oyr
- title :
- IPSL-CM6A-LR model output prepared for CMIP6 / CMIP piControl
- tracking_id :
- hdl:21.14100/5fb53c12-afe1-499d-9039-590b6e6b1501 hdl:21.14100/0825a99b-e57e-4796-9f3b-0d260c960c30
- variable_id :
- o2
- variant_info :
- Equivalent to r1i1p1f1 but started on a different machine. Information provided by this attribute may in some cases be flawed. Users can find more comprehensive and up-to-date documentation via the further_info_url global attribute.
- variant_label :
- r1i2p1f1
- status :
- 2019-10-25;created;by nhn2@columbia.edu
- netcdf_tracking_ids :
- hdl:21.14100/5fb53c12-afe1-499d-9039-590b6e6b1501 hdl:21.14100/0825a99b-e57e-4796-9f3b-0d260c960c30
- version_id :
- v20190319
- intake_esm_varname :
- ['o2']
- intake_esm_dataset_key :
- CMIP.IPSL.IPSL-CM6A-LR.piControl.Oyr.gn
[9]:
dset_dict_renamed.keys()
[9]:
dict_keys(['CMIP.IPSL.IPSL-CM6A-LR.piControl.Oyr.gn', 'CMIP.NOAA-GFDL.GFDL-ESM4.piControl.Oyr.gr', 'CMIP.CCCma.CanESM5.piControl.Oyr.gn'])
Nice. Now check out the GFDL model…
[10]:
ds = dset_dict_renamed['CMIP.NOAA-GFDL.GFDL-ESM4.piControl.Oyr.gr']
ds
[10]:
<xarray.Dataset>
Dimensions: (bnds: 2, lev: 35, member_id: 1, time: 500, x: 360, y: 180)
Coordinates:
* y (y) float64 -89.5 -88.5 -87.5 -86.5 ... 86.5 87.5 88.5 89.5
lat_bounds (y, bnds) float64 dask.array<chunksize=(180, 2), meta=np.ndarray>
* lev (lev) float64 2.5 10.0 20.0 32.5 ... 5.5e+03 6e+03 6.5e+03
lev_bounds (lev, bnds) float64 dask.array<chunksize=(35, 2), meta=np.ndarray>
* x (x) float64 0.5 1.5 2.5 3.5 4.5 ... 356.5 357.5 358.5 359.5
lon_bounds (x, bnds) float64 dask.array<chunksize=(360, 2), meta=np.ndarray>
* time (time) int64 0 365 730 1095 ... 181040 181405 181770 182135
time_bounds (time, bnds) float64 dask.array<chunksize=(500, 2), meta=np.ndarray>
* member_id (member_id) <U8 'r1i1p1f1'
Dimensions without coordinates: bnds
Data variables:
o2 (member_id, time, lev, y, x) float32 dask.array<chunksize=(1, 28, 35, 180, 360), meta=np.ndarray>
Attributes:
Conventions: CF-1.7 CMIP-6.0 UGRID-1.0
activity_id: CMIP
branch_method: Coupled climate state after 400 years of spinup,...
branch_time_in_child: 0.0
branch_time_in_parent: 0.0
comment: <null ref>
contact: gfdl.climate.model.info@noaa.gov
creation_date: 2019-05-13T21:46:48Z
data_specs_version: 01.00.27
experiment: pre-industrial control
experiment_id: piControl
external_variables: areacello volcello
forcing_index: 1
frequency: yr
further_info_url: https://furtherinfo.es-doc.org/CMIP6.NOAA-GFDL.G...
grid: ocean data regridded from tripolar - nominal 0.5...
grid_label: gr
history: File was processed by fremetar (GFDL analog of C...
initialization_index: 1
institution: National Oceanic and Atmospheric Administration,...
institution_id: NOAA-GFDL
license: CMIP6 model data produced by NOAA-GFDL is licens...
mip_era: CMIP6
nominal_resolution: 1x1 degree
parent_activity_id: CMIP
parent_experiment_id: piControl-spinup
parent_mip_era: CMIP6
parent_source_id: GFDL-ESM4
parent_time_units: days since 0001-1-1
parent_variant_label: r1i1p1f1
physics_index: 1
product: model-output
realization_index: 1
realm: ocnBgchem
references: see further_info_url attribute
source: GFDL-ESM4 (2018):\natmos: GFDL-AM4.1 (Cubed-sphe...
source_id: GFDL-ESM4
source_type: AOGCM AER CHEM BGC
sub_experiment: none
sub_experiment_id: none
table_id: Oyr
title: NOAA GFDL GFDL-ESM4 model output prepared for CM...
tracking_id: hdl:21.14100/894329be-9c3c-4dae-af32-3030d451e78...
variable_id: o2
variant_info: N/A
variant_label: r1i1p1f1
status: 2019-10-25;created;by nhn2@columbia.edu
netcdf_tracking_ids: hdl:21.14100/894329be-9c3c-4dae-af32-3030d451e78...
version_id: v20180701
intake_esm_varname: ['o2']
intake_esm_dataset_key: CMIP.NOAA-GFDL.GFDL-ESM4.piControl.Oyr.gr- bnds: 2
- lev: 35
- member_id: 1
- time: 500
- x: 360
- y: 180
- y(y)float64-89.5 -88.5 -87.5 ... 88.5 89.5
- axis :
- Y
- bounds :
- lat_bnds
- cell_methods :
- time: point
- long_name :
- latitude
- standard_name :
- latitude
- units :
- degrees_north
array([-89.5, -88.5, -87.5, -86.5, -85.5, -84.5, -83.5, -82.5, -81.5, -80.5, -79.5, -78.5, -77.5, -76.5, -75.5, -74.5, -73.5, -72.5, -71.5, -70.5, -69.5, -68.5, -67.5, -66.5, -65.5, -64.5, -63.5, -62.5, -61.5, -60.5, -59.5, -58.5, -57.5, -56.5, -55.5, -54.5, -53.5, -52.5, -51.5, -50.5, -49.5, -48.5, -47.5, -46.5, -45.5, -44.5, -43.5, -42.5, -41.5, -40.5, -39.5, -38.5, -37.5, -36.5, -35.5, -34.5, -33.5, -32.5, -31.5, -30.5, -29.5, -28.5, -27.5, -26.5, -25.5, -24.5, -23.5, -22.5, -21.5, -20.5, -19.5, -18.5, -17.5, -16.5, -15.5, -14.5, -13.5, -12.5, -11.5, -10.5, -9.5, -8.5, -7.5, -6.5, -5.5, -4.5, -3.5, -2.5, -1.5, -0.5, 0.5, 1.5, 2.5, 3.5, 4.5, 5.5, 6.5, 7.5, 8.5, 9.5, 10.5, 11.5, 12.5, 13.5, 14.5, 15.5, 16.5, 17.5, 18.5, 19.5, 20.5, 21.5, 22.5, 23.5, 24.5, 25.5, 26.5, 27.5, 28.5, 29.5, 30.5, 31.5, 32.5, 33.5, 34.5, 35.5, 36.5, 37.5, 38.5, 39.5, 40.5, 41.5, 42.5, 43.5, 44.5, 45.5, 46.5, 47.5, 48.5, 49.5, 50.5, 51.5, 52.5, 53.5, 54.5, 55.5, 56.5, 57.5, 58.5, 59.5, 60.5, 61.5, 62.5, 63.5, 64.5, 65.5, 66.5, 67.5, 68.5, 69.5, 70.5, 71.5, 72.5, 73.5, 74.5, 75.5, 76.5, 77.5, 78.5, 79.5, 80.5, 81.5, 82.5, 83.5, 84.5, 85.5, 86.5, 87.5, 88.5, 89.5]) - lat_bounds(y, bnds)float64dask.array<chunksize=(180, 2), meta=np.ndarray>
- axis :
- Y
- long_name :
- latitude bounds
- units :
- degrees_north
Array Chunk Bytes 2.88 kB 2.88 kB Shape (180, 2) (180, 2) Count 2 Tasks 1 Chunks Type float64 numpy.ndarray - lev(lev)float642.5 10.0 20.0 ... 6e+03 6.5e+03
- axis :
- Z
- bounds :
- lev_bnds
- description :
- generic ocean model vertical coordinate (nondimensional or dimensional)
- long_name :
- ocean model level
- positive :
- down
- units :
- m
array([2.5000e+00, 1.0000e+01, 2.0000e+01, 3.2500e+01, 5.1250e+01, 7.5000e+01, 1.0000e+02, 1.2500e+02, 1.5625e+02, 2.0000e+02, 2.5000e+02, 3.1250e+02, 4.0000e+02, 5.0000e+02, 6.0000e+02, 7.0000e+02, 8.0000e+02, 9.0000e+02, 1.0000e+03, 1.1000e+03, 1.2000e+03, 1.3000e+03, 1.4000e+03, 1.5375e+03, 1.7500e+03, 2.0625e+03, 2.5000e+03, 3.0000e+03, 3.5000e+03, 4.0000e+03, 4.5000e+03, 5.0000e+03, 5.5000e+03, 6.0000e+03, 6.5000e+03]) - lev_bounds(lev, bnds)float64dask.array<chunksize=(35, 2), meta=np.ndarray>
Array Chunk Bytes 560 B 560 B Shape (35, 2) (35, 2) Count 2 Tasks 1 Chunks Type float64 numpy.ndarray - x(x)float640.5 1.5 2.5 ... 357.5 358.5 359.5
- axis :
- X
- bounds :
- lon_bnds
- cell_methods :
- time: point
- long_name :
- longitude
- standard_name :
- longitude
- units :
- degrees_east
array([ 0.5, 1.5, 2.5, ..., 357.5, 358.5, 359.5])
- lon_bounds(x, bnds)float64dask.array<chunksize=(360, 2), meta=np.ndarray>
- axis :
- X
- long_name :
- longitude bounds
- units :
- degrees_east
Array Chunk Bytes 5.76 kB 5.76 kB Shape (360, 2) (360, 2) Count 2 Tasks 1 Chunks Type float64 numpy.ndarray - time(time)int640 365 730 ... 181405 181770 182135
- axis :
- T
- bounds :
- time_bnds
- calendar :
- noleap
- calendar_type :
- noleap
- description :
- Temporal mean
- long_name :
- time
- standard_name :
- time
- units :
- days since 0001-07-02 12:00:00.000000
array([ 0, 365, 730, ..., 181405, 181770, 182135])
- time_bounds(time, bnds)float64dask.array<chunksize=(500, 2), meta=np.ndarray>
- calendar :
- noleap
- long_name :
- time axis boundaries
- units :
- days since 0001-01-01
Array Chunk Bytes 8.00 kB 8.00 kB Shape (500, 2) (500, 2) Count 2 Tasks 1 Chunks Type float64 numpy.ndarray - member_id(member_id)<U8'r1i1p1f1'
array(['r1i1p1f1'], dtype='<U8')
- o2(member_id, time, lev, y, x)float32dask.array<chunksize=(1, 28, 35, 180, 360), meta=np.ndarray>
- cell_measures :
- area: areacello volume: volcello
- cell_methods :
- area: mean where sea time: mean
- comment :
- Model data on the 1x1 grid includes values in all cells for which any ocean exists on the native grid. For mapping purposes, we recommend using a land mask such as World Ocean Atlas to cover these areas of partial land. For calculating approximate integrals, we recommend multiplying by cell volume (volcello).
- interp_method :
- conserve_order1
- long_name :
- Dissolved Oxygen Concentration
- original_name :
- o2
- standard_name :
- mole_concentration_of_dissolved_molecular_oxygen_in_sea_water
- units :
- mol m-3
Array Chunk Bytes 4.54 GB 254.02 MB Shape (1, 500, 35, 180, 360) (1, 28, 35, 180, 360) Count 37 Tasks 18 Chunks Type float32 numpy.ndarray
- Conventions :
- CF-1.7 CMIP-6.0 UGRID-1.0
- activity_id :
- CMIP
- branch_method :
- Coupled climate state after 400 years of spinup, except land state after 800 years of spinup.
- branch_time_in_child :
- 0.0
- branch_time_in_parent :
- 0.0
- comment :
- <null ref>
- contact :
- gfdl.climate.model.info@noaa.gov
- creation_date :
- 2019-05-13T21:46:48Z
- data_specs_version :
- 01.00.27
- experiment :
- pre-industrial control
- experiment_id :
- piControl
- external_variables :
- areacello volcello
- forcing_index :
- 1
- frequency :
- yr
- further_info_url :
- https://furtherinfo.es-doc.org/CMIP6.NOAA-GFDL.GFDL-ESM4.piControl.none.r1i1p1f1
- grid :
- ocean data regridded from tripolar - nominal 0.5 deg latitude/longitude to 180,360; interpolation method: conserve_order1
- grid_label :
- gr
- history :
- File was processed by fremetar (GFDL analog of CMOR). TripleID: [exper_id_Nl8y6FcDL6,realiz_id_mNbn4PTB8f,run_id_0l1ykpVYGy]
- initialization_index :
- 1
- institution :
- National Oceanic and Atmospheric Administration, Geophysical Fluid Dynamics Laboratory, Princeton, NJ 08540, USA
- institution_id :
- NOAA-GFDL
- license :
- CMIP6 model data produced by NOAA-GFDL is licensed under a Creative Commons Attribution-ShareAlike 4.0 International License (https://creativecommons.org/licenses/). Consult https://pcmdi.llnl.gov/CMIP6/TermsOfUse for terms of use governing CMIP6 output, including citation requirements and proper acknowledgment. Further information about this data, including some limitations, can be found via the further_info_url (recorded as a global attribute in this file). The data producers and data providers make no warranty, either express or implied, including, but not limited to, warranties of merchantability and fitness for a particular purpose. All liabilities arising from the supply of the information (including any liability arising in negligence) are excluded to the fullest extent permitted by law.
- mip_era :
- CMIP6
- nominal_resolution :
- 1x1 degree
- parent_activity_id :
- CMIP
- parent_experiment_id :
- piControl-spinup
- parent_mip_era :
- CMIP6
- parent_source_id :
- GFDL-ESM4
- parent_time_units :
- days since 0001-1-1
- parent_variant_label :
- r1i1p1f1
- physics_index :
- 1
- product :
- model-output
- realization_index :
- 1
- realm :
- ocnBgchem
- references :
- see further_info_url attribute
- source :
- GFDL-ESM4 (2018): atmos: GFDL-AM4.1 (Cubed-sphere (c96) - 1 degree nominal horizontal resolution; 360 x 180 longitude/latitude; 49 levels; top level 1 Pa) ocean: GFDL-OM4p5 (GFDL-MOM6, tripolar - nominal 0.5 deg; 720 x 576 longitude/latitude; 75 levels; top grid cell 0-2 m) seaIce: GFDL-SIM4p5 (GFDL-SIS2.0, tripolar - nominal 0.5 deg; 720 x 576 longitude/latitude; 5 layers; 5 thickness categories) land: GFDL-LM4.1 aerosol: interactive atmosChem: GFDL-ATMCHEM4.1 (full atmospheric chemistry) ocnBgchem: GFDL-COBALTv2 landIce: GFDL-LM4.1 (GFDL ID: 2019_0273)
- source_id :
- GFDL-ESM4
- source_type :
- AOGCM AER CHEM BGC
- sub_experiment :
- none
- sub_experiment_id :
- none
- table_id :
- Oyr
- title :
- NOAA GFDL GFDL-ESM4 model output prepared for CMIP6 pre-industrial control
- tracking_id :
- hdl:21.14100/894329be-9c3c-4dae-af32-3030d451e789 hdl:21.14100/37a9d5ed-9ce3-4b7e-8524-75a6e9c962a6 hdl:21.14100/006ca735-4858-4ca3-8b5b-43a1735941a2 hdl:21.14100/9ded7d69-4429-4341-97ea-21f5588c42a4 hdl:21.14100/51554b9d-33ef-4358-b00b-404851a2cc97
- variable_id :
- o2
- variant_info :
- N/A
- variant_label :
- r1i1p1f1
- status :
- 2019-10-25;created;by nhn2@columbia.edu
- netcdf_tracking_ids :
- hdl:21.14100/894329be-9c3c-4dae-af32-3030d451e789 hdl:21.14100/37a9d5ed-9ce3-4b7e-8524-75a6e9c962a6 hdl:21.14100/006ca735-4858-4ca3-8b5b-43a1735941a2 hdl:21.14100/9ded7d69-4429-4341-97ea-21f5588c42a4 hdl:21.14100/51554b9d-33ef-4358-b00b-404851a2cc97
- version_id :
- v20180701
- intake_esm_varname :
- ['o2']
- intake_esm_dataset_key :
- CMIP.NOAA-GFDL.GFDL-ESM4.piControl.Oyr.gr
This dataset is from regridded output and has thus only 1D longitude and latitude values (which are called x and y due to the previous renaming step. broadcast_lonlat adds the lon and lat arrays back as 2d arrays.
[11]:
ds = broadcast_lonlat(ds)
ds
[11]:
<xarray.Dataset>
Dimensions: (bnds: 2, lev: 35, member_id: 1, time: 500, x: 360, y: 180)
Coordinates:
* y (y) float64 -89.5 -88.5 -87.5 -86.5 ... 86.5 87.5 88.5 89.5
lat_bounds (y, bnds) float64 dask.array<chunksize=(180, 2), meta=np.ndarray>
* lev (lev) float64 2.5 10.0 20.0 32.5 ... 5.5e+03 6e+03 6.5e+03
lev_bounds (lev, bnds) float64 dask.array<chunksize=(35, 2), meta=np.ndarray>
* x (x) float64 0.5 1.5 2.5 3.5 4.5 ... 356.5 357.5 358.5 359.5
lon_bounds (x, bnds) float64 dask.array<chunksize=(360, 2), meta=np.ndarray>
* time (time) int64 0 365 730 1095 ... 181040 181405 181770 182135
time_bounds (time, bnds) float64 dask.array<chunksize=(500, 2), meta=np.ndarray>
* member_id (member_id) <U8 'r1i1p1f1'
lon (x, y) float64 0.5 0.5 0.5 0.5 0.5 ... 359.5 359.5 359.5 359.5
lat (x, y) float64 -89.5 -88.5 -87.5 -86.5 ... 86.5 87.5 88.5 89.5
Dimensions without coordinates: bnds
Data variables:
o2 (member_id, time, lev, y, x) float32 dask.array<chunksize=(1, 28, 35, 180, 360), meta=np.ndarray>
Attributes:
Conventions: CF-1.7 CMIP-6.0 UGRID-1.0
activity_id: CMIP
branch_method: Coupled climate state after 400 years of spinup,...
branch_time_in_child: 0.0
branch_time_in_parent: 0.0
comment: <null ref>
contact: gfdl.climate.model.info@noaa.gov
creation_date: 2019-05-13T21:46:48Z
data_specs_version: 01.00.27
experiment: pre-industrial control
experiment_id: piControl
external_variables: areacello volcello
forcing_index: 1
frequency: yr
further_info_url: https://furtherinfo.es-doc.org/CMIP6.NOAA-GFDL.G...
grid: ocean data regridded from tripolar - nominal 0.5...
grid_label: gr
history: File was processed by fremetar (GFDL analog of C...
initialization_index: 1
institution: National Oceanic and Atmospheric Administration,...
institution_id: NOAA-GFDL
license: CMIP6 model data produced by NOAA-GFDL is licens...
mip_era: CMIP6
nominal_resolution: 1x1 degree
parent_activity_id: CMIP
parent_experiment_id: piControl-spinup
parent_mip_era: CMIP6
parent_source_id: GFDL-ESM4
parent_time_units: days since 0001-1-1
parent_variant_label: r1i1p1f1
physics_index: 1
product: model-output
realization_index: 1
realm: ocnBgchem
references: see further_info_url attribute
source: GFDL-ESM4 (2018):\natmos: GFDL-AM4.1 (Cubed-sphe...
source_id: GFDL-ESM4
source_type: AOGCM AER CHEM BGC
sub_experiment: none
sub_experiment_id: none
table_id: Oyr
title: NOAA GFDL GFDL-ESM4 model output prepared for CM...
tracking_id: hdl:21.14100/894329be-9c3c-4dae-af32-3030d451e78...
variable_id: o2
variant_info: N/A
variant_label: r1i1p1f1
status: 2019-10-25;created;by nhn2@columbia.edu
netcdf_tracking_ids: hdl:21.14100/894329be-9c3c-4dae-af32-3030d451e78...
version_id: v20180701
intake_esm_varname: ['o2']
intake_esm_dataset_key: CMIP.NOAA-GFDL.GFDL-ESM4.piControl.Oyr.gr- bnds: 2
- lev: 35
- member_id: 1
- time: 500
- x: 360
- y: 180
- y(y)float64-89.5 -88.5 -87.5 ... 88.5 89.5
- axis :
- Y
- bounds :
- lat_bnds
- cell_methods :
- time: point
- long_name :
- latitude
- standard_name :
- latitude
- units :
- degrees_north
array([-89.5, -88.5, -87.5, -86.5, -85.5, -84.5, -83.5, -82.5, -81.5, -80.5, -79.5, -78.5, -77.5, -76.5, -75.5, -74.5, -73.5, -72.5, -71.5, -70.5, -69.5, -68.5, -67.5, -66.5, -65.5, -64.5, -63.5, -62.5, -61.5, -60.5, -59.5, -58.5, -57.5, -56.5, -55.5, -54.5, -53.5, -52.5, -51.5, -50.5, -49.5, -48.5, -47.5, -46.5, -45.5, -44.5, -43.5, -42.5, -41.5, -40.5, -39.5, -38.5, -37.5, -36.5, -35.5, -34.5, -33.5, -32.5, -31.5, -30.5, -29.5, -28.5, -27.5, -26.5, -25.5, -24.5, -23.5, -22.5, -21.5, -20.5, -19.5, -18.5, -17.5, -16.5, -15.5, -14.5, -13.5, -12.5, -11.5, -10.5, -9.5, -8.5, -7.5, -6.5, -5.5, -4.5, -3.5, -2.5, -1.5, -0.5, 0.5, 1.5, 2.5, 3.5, 4.5, 5.5, 6.5, 7.5, 8.5, 9.5, 10.5, 11.5, 12.5, 13.5, 14.5, 15.5, 16.5, 17.5, 18.5, 19.5, 20.5, 21.5, 22.5, 23.5, 24.5, 25.5, 26.5, 27.5, 28.5, 29.5, 30.5, 31.5, 32.5, 33.5, 34.5, 35.5, 36.5, 37.5, 38.5, 39.5, 40.5, 41.5, 42.5, 43.5, 44.5, 45.5, 46.5, 47.5, 48.5, 49.5, 50.5, 51.5, 52.5, 53.5, 54.5, 55.5, 56.5, 57.5, 58.5, 59.5, 60.5, 61.5, 62.5, 63.5, 64.5, 65.5, 66.5, 67.5, 68.5, 69.5, 70.5, 71.5, 72.5, 73.5, 74.5, 75.5, 76.5, 77.5, 78.5, 79.5, 80.5, 81.5, 82.5, 83.5, 84.5, 85.5, 86.5, 87.5, 88.5, 89.5]) - lat_bounds(y, bnds)float64dask.array<chunksize=(180, 2), meta=np.ndarray>
- axis :
- Y
- long_name :
- latitude bounds
- units :
- degrees_north
Array Chunk Bytes 2.88 kB 2.88 kB Shape (180, 2) (180, 2) Count 2 Tasks 1 Chunks Type float64 numpy.ndarray - lev(lev)float642.5 10.0 20.0 ... 6e+03 6.5e+03
- axis :
- Z
- bounds :
- lev_bnds
- description :
- generic ocean model vertical coordinate (nondimensional or dimensional)
- long_name :
- ocean model level
- positive :
- down
- units :
- m
array([2.5000e+00, 1.0000e+01, 2.0000e+01, 3.2500e+01, 5.1250e+01, 7.5000e+01, 1.0000e+02, 1.2500e+02, 1.5625e+02, 2.0000e+02, 2.5000e+02, 3.1250e+02, 4.0000e+02, 5.0000e+02, 6.0000e+02, 7.0000e+02, 8.0000e+02, 9.0000e+02, 1.0000e+03, 1.1000e+03, 1.2000e+03, 1.3000e+03, 1.4000e+03, 1.5375e+03, 1.7500e+03, 2.0625e+03, 2.5000e+03, 3.0000e+03, 3.5000e+03, 4.0000e+03, 4.5000e+03, 5.0000e+03, 5.5000e+03, 6.0000e+03, 6.5000e+03]) - lev_bounds(lev, bnds)float64dask.array<chunksize=(35, 2), meta=np.ndarray>
Array Chunk Bytes 560 B 560 B Shape (35, 2) (35, 2) Count 2 Tasks 1 Chunks Type float64 numpy.ndarray - x(x)float640.5 1.5 2.5 ... 357.5 358.5 359.5
- axis :
- X
- bounds :
- lon_bnds
- cell_methods :
- time: point
- long_name :
- longitude
- standard_name :
- longitude
- units :
- degrees_east
array([ 0.5, 1.5, 2.5, ..., 357.5, 358.5, 359.5])
- lon_bounds(x, bnds)float64dask.array<chunksize=(360, 2), meta=np.ndarray>
- axis :
- X
- long_name :
- longitude bounds
- units :
- degrees_east
Array Chunk Bytes 5.76 kB 5.76 kB Shape (360, 2) (360, 2) Count 2 Tasks 1 Chunks Type float64 numpy.ndarray - time(time)int640 365 730 ... 181405 181770 182135
- axis :
- T
- bounds :
- time_bnds
- calendar :
- noleap
- calendar_type :
- noleap
- description :
- Temporal mean
- long_name :
- time
- standard_name :
- time
- units :
- days since 0001-07-02 12:00:00.000000
array([ 0, 365, 730, ..., 181405, 181770, 182135])
- time_bounds(time, bnds)float64dask.array<chunksize=(500, 2), meta=np.ndarray>
- calendar :
- noleap
- long_name :
- time axis boundaries
- units :
- days since 0001-01-01
Array Chunk Bytes 8.00 kB 8.00 kB Shape (500, 2) (500, 2) Count 2 Tasks 1 Chunks Type float64 numpy.ndarray - member_id(member_id)<U8'r1i1p1f1'
array(['r1i1p1f1'], dtype='<U8')
- lon(x, y)float640.5 0.5 0.5 ... 359.5 359.5 359.5
array([[ 0.5, 0.5, 0.5, ..., 0.5, 0.5, 0.5], [ 1.5, 1.5, 1.5, ..., 1.5, 1.5, 1.5], [ 2.5, 2.5, 2.5, ..., 2.5, 2.5, 2.5], ..., [357.5, 357.5, 357.5, ..., 357.5, 357.5, 357.5], [358.5, 358.5, 358.5, ..., 358.5, 358.5, 358.5], [359.5, 359.5, 359.5, ..., 359.5, 359.5, 359.5]]) - lat(x, y)float64-89.5 -88.5 -87.5 ... 88.5 89.5
array([[-89.5, -88.5, -87.5, ..., 87.5, 88.5, 89.5], [-89.5, -88.5, -87.5, ..., 87.5, 88.5, 89.5], [-89.5, -88.5, -87.5, ..., 87.5, 88.5, 89.5], ..., [-89.5, -88.5, -87.5, ..., 87.5, 88.5, 89.5], [-89.5, -88.5, -87.5, ..., 87.5, 88.5, 89.5], [-89.5, -88.5, -87.5, ..., 87.5, 88.5, 89.5]])
- o2(member_id, time, lev, y, x)float32dask.array<chunksize=(1, 28, 35, 180, 360), meta=np.ndarray>
- cell_measures :
- area: areacello volume: volcello
- cell_methods :
- area: mean where sea time: mean
- comment :
- Model data on the 1x1 grid includes values in all cells for which any ocean exists on the native grid. For mapping purposes, we recommend using a land mask such as World Ocean Atlas to cover these areas of partial land. For calculating approximate integrals, we recommend multiplying by cell volume (volcello).
- interp_method :
- conserve_order1
- long_name :
- Dissolved Oxygen Concentration
- original_name :
- o2
- standard_name :
- mole_concentration_of_dissolved_molecular_oxygen_in_sea_water
- units :
- mol m-3
Array Chunk Bytes 4.54 GB 254.02 MB Shape (1, 500, 35, 180, 360) (1, 28, 35, 180, 360) Count 37 Tasks 18 Chunks Type float32 numpy.ndarray
- Conventions :
- CF-1.7 CMIP-6.0 UGRID-1.0
- activity_id :
- CMIP
- branch_method :
- Coupled climate state after 400 years of spinup, except land state after 800 years of spinup.
- branch_time_in_child :
- 0.0
- branch_time_in_parent :
- 0.0
- comment :
- <null ref>
- contact :
- gfdl.climate.model.info@noaa.gov
- creation_date :
- 2019-05-13T21:46:48Z
- data_specs_version :
- 01.00.27
- experiment :
- pre-industrial control
- experiment_id :
- piControl
- external_variables :
- areacello volcello
- forcing_index :
- 1
- frequency :
- yr
- further_info_url :
- https://furtherinfo.es-doc.org/CMIP6.NOAA-GFDL.GFDL-ESM4.piControl.none.r1i1p1f1
- grid :
- ocean data regridded from tripolar - nominal 0.5 deg latitude/longitude to 180,360; interpolation method: conserve_order1
- grid_label :
- gr
- history :
- File was processed by fremetar (GFDL analog of CMOR). TripleID: [exper_id_Nl8y6FcDL6,realiz_id_mNbn4PTB8f,run_id_0l1ykpVYGy]
- initialization_index :
- 1
- institution :
- National Oceanic and Atmospheric Administration, Geophysical Fluid Dynamics Laboratory, Princeton, NJ 08540, USA
- institution_id :
- NOAA-GFDL
- license :
- CMIP6 model data produced by NOAA-GFDL is licensed under a Creative Commons Attribution-ShareAlike 4.0 International License (https://creativecommons.org/licenses/). Consult https://pcmdi.llnl.gov/CMIP6/TermsOfUse for terms of use governing CMIP6 output, including citation requirements and proper acknowledgment. Further information about this data, including some limitations, can be found via the further_info_url (recorded as a global attribute in this file). The data producers and data providers make no warranty, either express or implied, including, but not limited to, warranties of merchantability and fitness for a particular purpose. All liabilities arising from the supply of the information (including any liability arising in negligence) are excluded to the fullest extent permitted by law.
- mip_era :
- CMIP6
- nominal_resolution :
- 1x1 degree
- parent_activity_id :
- CMIP
- parent_experiment_id :
- piControl-spinup
- parent_mip_era :
- CMIP6
- parent_source_id :
- GFDL-ESM4
- parent_time_units :
- days since 0001-1-1
- parent_variant_label :
- r1i1p1f1
- physics_index :
- 1
- product :
- model-output
- realization_index :
- 1
- realm :
- ocnBgchem
- references :
- see further_info_url attribute
- source :
- GFDL-ESM4 (2018): atmos: GFDL-AM4.1 (Cubed-sphere (c96) - 1 degree nominal horizontal resolution; 360 x 180 longitude/latitude; 49 levels; top level 1 Pa) ocean: GFDL-OM4p5 (GFDL-MOM6, tripolar - nominal 0.5 deg; 720 x 576 longitude/latitude; 75 levels; top grid cell 0-2 m) seaIce: GFDL-SIM4p5 (GFDL-SIS2.0, tripolar - nominal 0.5 deg; 720 x 576 longitude/latitude; 5 layers; 5 thickness categories) land: GFDL-LM4.1 aerosol: interactive atmosChem: GFDL-ATMCHEM4.1 (full atmospheric chemistry) ocnBgchem: GFDL-COBALTv2 landIce: GFDL-LM4.1 (GFDL ID: 2019_0273)
- source_id :
- GFDL-ESM4
- source_type :
- AOGCM AER CHEM BGC
- sub_experiment :
- none
- sub_experiment_id :
- none
- table_id :
- Oyr
- title :
- NOAA GFDL GFDL-ESM4 model output prepared for CMIP6 pre-industrial control
- tracking_id :
- hdl:21.14100/894329be-9c3c-4dae-af32-3030d451e789 hdl:21.14100/37a9d5ed-9ce3-4b7e-8524-75a6e9c962a6 hdl:21.14100/006ca735-4858-4ca3-8b5b-43a1735941a2 hdl:21.14100/9ded7d69-4429-4341-97ea-21f5588c42a4 hdl:21.14100/51554b9d-33ef-4358-b00b-404851a2cc97
- variable_id :
- o2
- variant_info :
- N/A
- variant_label :
- r1i1p1f1
- status :
- 2019-10-25;created;by nhn2@columbia.edu
- netcdf_tracking_ids :
- hdl:21.14100/894329be-9c3c-4dae-af32-3030d451e789 hdl:21.14100/37a9d5ed-9ce3-4b7e-8524-75a6e9c962a6 hdl:21.14100/006ca735-4858-4ca3-8b5b-43a1735941a2 hdl:21.14100/9ded7d69-4429-4341-97ea-21f5588c42a4 hdl:21.14100/51554b9d-33ef-4358-b00b-404851a2cc97
- version_id :
- v20180701
- intake_esm_varname :
- ['o2']
- intake_esm_dataset_key :
- CMIP.NOAA-GFDL.GFDL-ESM4.piControl.Oyr.gr
When you look back at the IPSL model you notice that the x and y values are given just as indicies, making rough selection of regions using xarrays .sel rather useless. To gain back this functionality, we replace x and y with nominal longitudes and latitudes using replace_x_y_nominal_lat_lon:
As of version ‘0.2’ this is not part of the
combined_preprocessinganymore due to performance issue, when applied to a full dataset. You can still apply this function before plotting as shown in the final part of the tutorial
[12]:
ds = dset_dict_renamed['CMIP.CCCma.CanESM5.piControl.Oyr.gn']
print(ds.y.data)
ds = replace_x_y_nominal_lat_lon(ds)
ds.y.data
[ 0 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17
18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35
36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 52 53
54 55 56 57 58 59 60 61 62 63 64 65 66 67 68 69 70 71
72 73 74 75 76 77 78 79 80 81 82 83 84 85 86 87 88 89
90 91 92 93 94 95 96 97 98 99 100 101 102 103 104 105 106 107
108 109 110 111 112 113 114 115 116 117 118 119 120 121 122 123 124 125
126 127 128 129 130 131 132 133 134 135 136 137 138 139 140 141 142 143
144 145 146 147 148 149 150 151 152 153 154 155 156 157 158 159 160 161
162 163 164 165 166 167 168 169 170 171 172 173 174 175 176 177 178 179
180 181 182 183 184 185 186 187 188 189 190 191 192 193 194 195 196 197
198 199 200 201 202 203 204 205 206 207 208 209 210 211 212 213 214 215
216 217 218 219 220 221 222 223 224 225 226 227 228 229 230 231 232 233
234 235 236 237 238 239 240 241 242 243 244 245 246 247 248 249 250 251
252 253 254 255 256 257 258 259 260 261 262 263 264 265 266 267 268 269
270 271 272 273 274 275 276 277 278 279 280 281 282 283 284 285 286 287
288 289 290]
/Users/juliusbusecke/miniconda/envs/xmip_docs/lib/python3.9/site-packages/xarray/core/indexing.py:1369: PerformanceWarning: Slicing is producing a large chunk. To accept the large
chunk and silence this warning, set the option
>>> with dask.config.set(**{'array.slicing.split_large_chunks': False}):
... array[indexer]
To avoid creating the large chunks, set the option
>>> with dask.config.set(**{'array.slicing.split_large_chunks': True}):
... array[indexer]
return self.array[key]
[12]:
array([-78.39350128, -78.19058228, -77.98416901, -77.77420044,
-77.56062317, -77.34336853, -77.12238312, -76.89761353,
-76.66898346, -76.43643951, -76.19991302, -75.95934296,
-75.71466827, -75.46582031, -75.21273041, -74.95533752,
-74.69356537, -74.42735291, -74.15662384, -73.88130951,
-73.60134125, -73.31665039, -73.02715302, -72.73278809,
-72.43347168, -72.12913513, -71.81970215, -71.50509644,
-71.18523407, -70.86004639, -70.52945709, -70.19337463,
-69.85173798, -69.50444794, -69.15143585, -68.79262543,
-68.42792511, -68.05725098, -67.68053436, -67.29768372,
-66.90862274, -66.51325989, -66.11151886, -65.70331573,
-65.28856659, -64.86719513, -64.43910217, -64.00422668,
-63.56246948, -63.11375427, -62.65800095, -62.19512558,
-61.72504807, -61.24769211, -60.76297379, -60.27082062,
-59.77114868, -59.2638855 , -58.74895477, -58.22628403,
-57.69579697, -57.15742874, -56.61111069, -56.05677032,
-55.49434662, -54.92377472, -54.34499359, -53.75794983,
-53.1625824 , -52.55883789, -51.94667435, -51.32603455,
-50.69688416, -50.0591774 , -49.41287994, -48.75795746,
-48.09438705, -47.42214203, -46.74119949, -46.051548 ,
-45.35317612, -44.6460762 , -43.93025208, -43.20571136,
-42.47245789, -41.73051071, -40.97989655, -40.22064209,
-39.45278549, -38.67636108, -37.89142609, -37.09803009,
-36.29623795, -35.48611832, -34.66775131, -33.84122086,
-33.00661469, -32.16403961, -31.31359863, -30.4554081 ,
-29.58959389, -28.7162838 , -27.83562279, -26.94775391,
-26.05283546, -25.15103149, -24.24251175, -23.32746124,
-22.40606308, -21.47851562, -20.54502296, -19.605793 ,
-18.66210938, -17.71509743, -16.76747131, -15.82362843,
-14.88963032, -13.97249222, -13.07900047, -12.21489906,
-11.3851366 , -10.59480476, -9.84941292, -9.1537075 ,
-8.50995922, -7.91714287, -7.3714118 , -6.86724281,
-6.39854765, -5.95941591, -5.54450321, -5.1491704 ,
-4.76948118, -4.40214634, -4.04443884, -3.69411659,
-3.3493495 , -3.00865722, -2.67085862, -2.33502841,
-2.00046086, -1.66663659, -1.33319354, -0.99990022,
-0.66662848, -0.33332792, 0. , 0.33332792,
0.66662848, 0.99990022, 1.33319354, 1.66663659,
2.00046086, 2.33502841, 2.67085862, 3.00865722,
3.3493495 , 3.69411659, 4.04443884, 4.40214634,
4.76948118, 5.1491704 , 5.54450321, 5.95941591,
6.39854765, 6.86724281, 7.3714118 , 7.91714287,
8.50995922, 9.1537075 , 9.84941292, 10.59480476,
11.3851366 , 12.21489906, 13.07900047, 13.97249222,
14.88963032, 15.82362843, 16.76747131, 17.71509743,
18.66210938, 19.605793 , 20.54502678, 21.4785862 ,
22.40633011, 23.32811546, 24.24381447, 25.15330124,
26.05646706, 26.95320511, 27.84342003, 28.72702217,
29.60393143, 30.4740715 , 31.33737755, 32.19379044,
33.04325867, 33.88573456, 34.72118759, 35.54957962,
36.37088776, 37.18510056, 37.99220276, 38.79219818,
39.58508682, 40.37087631, 41.14958954, 41.92126465,
42.68597031, 43.44370651, 44.19451523, 44.93845367,
45.67557907, 46.40594864, 47.12963867, 47.84671783,
48.55727386, 49.26138687, 49.95915222, 50.65066147,
51.33601761, 52.01532364, 52.68869019, 53.35645294,
54.01857376, 54.67513275, 55.3262558 , 55.97206116,
56.61268616, 57.24825668, 57.87890625, 58.50476456,
59.12596893, 59.74265289, 60.35513306, 60.96369171,
61.56817245, 62.16870499, 62.76543045, 63.35847473,
63.94796753, 64.53403473, 65.11679077, 65.69636536,
66.2728653 , 66.84711456, 67.41855621, 67.9872818 ,
68.55338287, 69.11695099, 69.67805481, 70.23677063,
70.79315948, 71.34726715, 71.89914703, 72.44923401,
72.99770355, 73.54405212, 74.08827972, 74.63038635,
75.17033386, 75.70809174, 76.24359894, 76.77680969,
77.30763245, 77.83597565, 78.36173248, 78.88496399,
79.40639496, 79.92492676, 80.44039154, 80.95262909,
81.46146393, 81.96670532, 82.46815491, 82.96560669,
83.45883179, 83.94761658, 84.43172455, 84.91091156,
85.38493347, 85.85354614, 86.31648254, 86.77348328,
87.22425842, 87.66850281, 88.10582733, 88.53568268,
88.95700073, 89.36653137, 89.74176788])
We can put all of this together in a wrapper function and plot some data
[13]:
def wrapper(ds):
ds = ds.copy()
ds = rename_cmip6(ds)
ds = promote_empty_dims(ds)
ds = broadcast_lonlat(ds)
ds = replace_x_y_nominal_lat_lon(ds)
return ds
# pass the preprocessing directly
with dask.config.set(**{'array.slicing.split_large_chunks': True}):
dset_dict_processed1 = cat.to_dataset_dict(zarr_kwargs=z_kwargs,
preprocess=wrapper)
--> The keys in the returned dictionary of datasets are constructed as follows:
'activity_id.institution_id.source_id.experiment_id.table_id.grid_label'
/Users/juliusbusecke/miniconda/envs/xmip_docs/lib/python3.9/site-packages/dask/array/core.py:4241: PerformanceWarning: Increasing number of chunks by factor of 60
result = blockwise(
[14]:
fig, axarr = plt.subplots(nrows=3, figsize=[10,15])
for ax, (k, ds) in zip(axarr.flat, dset_dict_processed1.items()):
if 'member_id' in ds.dims:
ds = ds.isel(member_id=-1)
ds.o2.isel(time=0, lev=0).sel(y=slice(-15,15)).plot(ax=ax)
ax.set_title(k)
ax.set_aspect(2)

The naming and units are still inconsistent (not implemented yet) and the longitude is not consistent (we will deal with this below) But this is a big step forward. With the ‘unprocessed’ datasets this would have needed a lot more logic in the print loop.
## Inconsistent longitude conventions We saw above that not all models have a ‘0-360’ longitude convention. We can fix this very quickly using correct_lon:
[15]:
from xmip.preprocessing import correct_lon
# same as above
def wrapper(ds):
ds = ds.copy()
ds = rename_cmip6(ds)
ds = promote_empty_dims(ds)
ds = broadcast_lonlat(ds)
ds = correct_lon(ds)
ds = replace_x_y_nominal_lat_lon(ds)
return ds
# pass the preprocessing directly
with dask.config.set(**{'array.slicing.split_large_chunks': True}):
dset_dict_processed2 = cat.to_dataset_dict(zarr_kwargs=z_kwargs,
preprocess=wrapper)
--> The keys in the returned dictionary of datasets are constructed as follows:
'activity_id.institution_id.source_id.experiment_id.table_id.grid_label'
/Users/juliusbusecke/miniconda/envs/xmip_docs/lib/python3.9/site-packages/dask/array/core.py:4241: PerformanceWarning: Increasing number of chunks by factor of 60
result = blockwise(
[16]:
fig, axarr = plt.subplots(nrows=3, figsize=[10,15])
for ax, (k, ds) in zip(axarr.flat, dset_dict_processed2.items()):
if 'member_id' in ds.dims:
ds = ds.isel(member_id=-1)
ds.o2.isel(time=0, lev=0).sel(y=slice(-15,15)).plot(ax=ax)
ax.set_title(k)
ax.set_aspect(2)
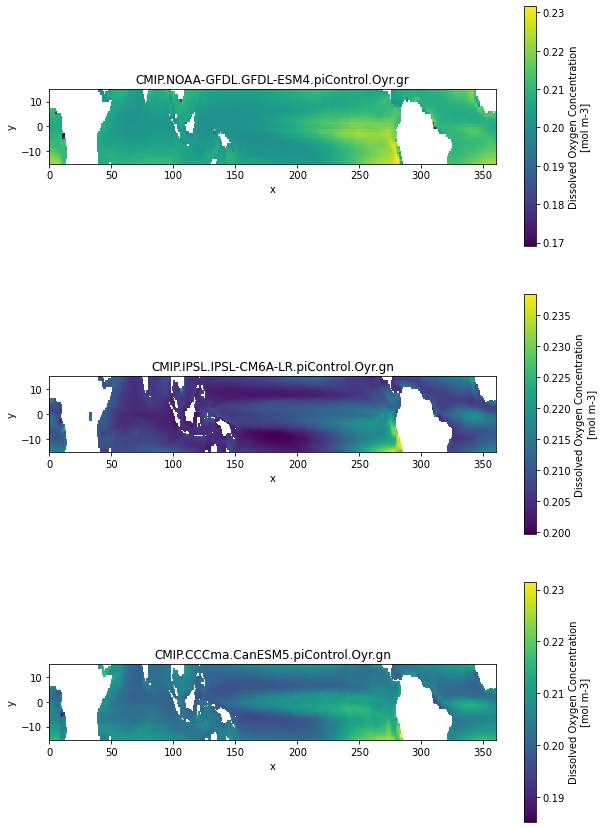

## Inconsistent units
But of course this is not all. Some models, give the depth in centimeters (so far I have only seen this in the NCAR models). We can fix this with correct_units:
[17]:
from xmip.preprocessing import correct_units
query = dict(experiment_id = ['historical'],variable_id='thetao', grid_label=['gn'],source_id=['CESM2', 'CanESM5'], member_id='r1i1p1f1',
)
cat = col.search(**query)
# raw data read in
dset_dict = cat.to_dataset_dict(zarr_kwargs=z_kwargs)
# fixed units
dset_dict_fixed_unit = cat.to_dataset_dict(zarr_kwargs=z_kwargs, preprocess=correct_units)
--> The keys in the returned dictionary of datasets are constructed as follows:
'activity_id.institution_id.source_id.experiment_id.table_id.grid_label'
--> The keys in the returned dictionary of datasets are constructed as follows:
'activity_id.institution_id.source_id.experiment_id.table_id.grid_label'
[18]:
dset_dict['CMIP.NCAR.CESM2.historical.Omon.gn'].lev.plot()
plt.figure()
dset_dict_fixed_unit['CMIP.NCAR.CESM2.historical.Omon.gn'].lev.plot()
[18]:
[<matplotlib.lines.Line2D at 0x170297640>]
This helps tremendously when you are trying to slice a common depth from a series of models
[19]:
fig, axarr = plt.subplots(nrows=2, figsize=[10,10])
for ax, (k, ds) in zip(axarr.flat, dset_dict_fixed_unit.items()):
ds.thetao.isel(time=0).sel(lev=5000, method='nearest').plot(ax=ax, vmin=-1, vmax=5)
ax.set_title(k)
As a comparison, for the unprocessed data this would have picked the depth at 50m for the CESM2 model instead of 5000m:
[20]:
fig, axarr = plt.subplots(nrows=2, figsize=[10,10])
for ax, (k, ds) in zip(axarr.flat, dset_dict.items()):
ds.thetao.isel(time=0).sel(lev=5000, method='nearest').plot(ax=ax, vmin=-1, vmax=5)
ax.set_title(k)
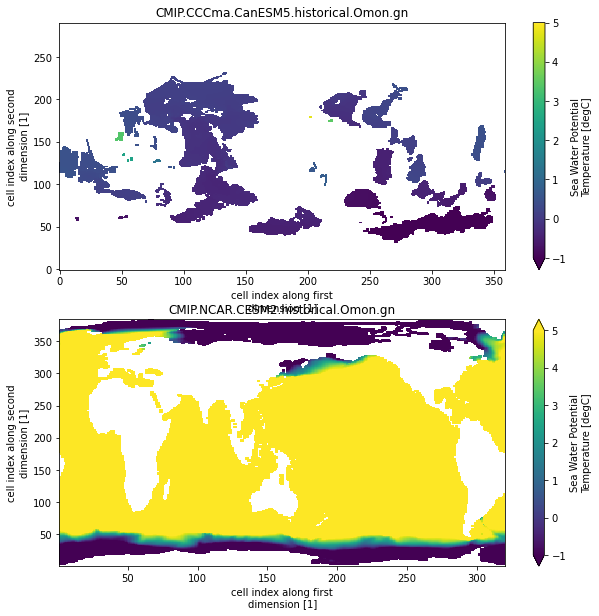
## Consistent CF bounds Many of the CMIP6 models come with ‘bound’ dataarrays, that describe the extent of the finite grid cells. For the longitude and latitude there are two conventions: 2-element ‘bounds’ (describing the width of a cell along the center) and 4 element ‘verticies’ (describing the 4 corner coordinates of the cell). xmip automatically renames these variables consistently and converts them so that every dataset has both conventions available.
[21]:
from xmip.preprocessing import correct_coordinates,parse_lon_lat_bounds, maybe_convert_bounds_to_vertex, maybe_convert_vertex_to_bounds
# same as above
def wrapper(ds):
ds = ds.copy()
ds = rename_cmip6(ds)
ds = promote_empty_dims(ds)
ds = broadcast_lonlat(ds)
ds = replace_x_y_nominal_lat_lon(ds)
ds = correct_lon(ds)
ds = correct_coordinates(ds)
ds = parse_lon_lat_bounds(ds)
ds = maybe_convert_bounds_to_vertex(ds)
ds = maybe_convert_vertex_to_bounds(ds)
return ds
# pass the preprocessing directly
dset_dict_processed3 = cat.to_dataset_dict(zarr_kwargs=z_kwargs,
preprocess=wrapper)
--> The keys in the returned dictionary of datasets are constructed as follows:
'activity_id.institution_id.source_id.experiment_id.table_id.grid_label'
/Users/juliusbusecke/miniconda/envs/xmip_docs/lib/python3.9/site-packages/xarray/core/indexing.py:1369: PerformanceWarning: Slicing is producing a large chunk. To accept the large
chunk and silence this warning, set the option
>>> with dask.config.set(**{'array.slicing.split_large_chunks': False}):
... array[indexer]
To avoid creating the large chunks, set the option
>>> with dask.config.set(**{'array.slicing.split_large_chunks': True}):
... array[indexer]
return self.array[key]
/Users/juliusbusecke/miniconda/envs/xmip_docs/lib/python3.9/site-packages/xarray/core/indexing.py:1369: PerformanceWarning: Slicing is producing a large chunk. To accept the large
chunk and silence this warning, set the option
>>> with dask.config.set(**{'array.slicing.split_large_chunks': False}):
... array[indexer]
To avoid creating the large chunks, set the option
>>> with dask.config.set(**{'array.slicing.split_large_chunks': True}):
... array[indexer]
return self.array[key]
/Users/juliusbusecke/miniconda/envs/xmip_docs/lib/python3.9/site-packages/xarray/core/indexing.py:1369: PerformanceWarning: Slicing is producing a large chunk. To accept the large
chunk and silence this warning, set the option
>>> with dask.config.set(**{'array.slicing.split_large_chunks': False}):
... array[indexer]
To avoid creating the large chunks, set the option
>>> with dask.config.set(**{'array.slicing.split_large_chunks': True}):
... array[indexer]
return self.array[key]
[22]:
for k, ds in dset_dict_processed3.items():
print(ds)
<xarray.Dataset>
Dimensions: (bnds: 2, lev: 60, member_id: 1, time: 1980, vertex: 4, x: 320, y: 384)
Coordinates:
lat (y, x) float64 dask.array<chunksize=(384, 320), meta=np.ndarray>
lat_verticies (y, x, vertex) float32 dask.array<chunksize=(384, 320, 4), meta=np.ndarray>
* lev (lev) float64 500.0 1.5e+03 2.5e+03 ... 5.125e+05 5.375e+05
lev_bounds (lev, bnds) float32 dask.array<chunksize=(60, 2), meta=np.ndarray>
lon (y, x) float64 dask.array<chunksize=(384, 320), meta=np.ndarray>
lon_verticies (y, x, vertex) float32 dask.array<chunksize=(384, 320, 4), meta=np.ndarray>
* y (y) float64 -79.22 -78.69 -78.15 -77.62 ... 89.11 89.66 89.71
* x (x) float64 1.062 2.187 3.312 4.437 ... 357.7 358.8 359.9
* time (time) float64 6.749e+05 6.749e+05 ... 7.351e+05 7.351e+05
time_bounds (time, bnds) float64 dask.array<chunksize=(1980, 2), meta=np.ndarray>
* bnds (bnds) int64 0 1
* vertex (vertex) int64 0 1 2 3
lon_bounds (bnds, y, x) float32 dask.array<chunksize=(1, 384, 320), meta=np.ndarray>
lat_bounds (bnds, y, x) float32 dask.array<chunksize=(1, 384, 320), meta=np.ndarray>
* member_id (member_id) <U8 'r1i1p1f1'
Data variables:
thetao (member_id, time, lev, y, x) float32 dask.array<chunksize=(1, 8, 60, 384, 320), meta=np.ndarray>
Attributes:
Conventions: CF-1.7 CMIP-6.2
activity_id: CMIP
branch_method: standard
branch_time_in_child: 674885.0
branch_time_in_parent: 219000.0
case_id: 15
cesm_casename: b.e21.BHIST.f09_g17.CMIP6-historical.001
contact: cesm_cmip6@ucar.edu
creation_date: 2019-01-16T22:01:19Z
data_specs_version: 01.00.29
experiment: all-forcing simulation of the recent past
experiment_id: historical
external_variables: areacello volcello
forcing_index: 1
frequency: mon
further_info_url: https://furtherinfo.es-doc.org/CMIP6.NCAR.CESM2....
grid: native gx1v7 displaced pole grid (384x320 latxlon)
grid_label: gn
initialization_index: 1
institution: National Center for Atmospheric Research, Climat...
institution_id: NCAR
license: CMIP6 model data produced by <The National Cente...
mip_era: CMIP6
model_doi_url: https://doi.org/10.5065/D67H1H0V
nominal_resolution: 100 km
parent_activity_id: CMIP
parent_experiment_id: piControl
parent_mip_era: CMIP6
parent_source_id: CESM2
parent_time_units: days since 0001-01-01 00:00:00
parent_variant_label: r1i1p1f1
physics_index: 1
product: model-output
realization_index: 1
realm: ocean
source: CESM2 (2017): atmosphere: CAM6 (0.9x1.25 finite ...
source_id: CESM2
source_type: AOGCM BGC
sub_experiment: none
sub_experiment_id: none
table_id: Omon
tracking_id: hdl:21.14100/672d35cd-e662-4807-8dee-7d7d5e1d4d1c
variable_id: thetao
variant_info: CMIP6 20th century experiments (1850-2014) with ...
variant_label: r1i1p1f1
status: 2019-10-25;created;by nhn2@columbia.edu
netcdf_tracking_ids: hdl:21.14100/672d35cd-e662-4807-8dee-7d7d5e1d4d1c
version_id: v20190308
intake_esm_varname: ['thetao']
intake_esm_dataset_key: CMIP.NCAR.CESM2.historical.Omon.gn
<xarray.Dataset>
Dimensions: (bnds: 2, lev: 45, member_id: 1, time: 1980, vertex: 4, x: 360, y: 291)
Coordinates:
* x (x) float64 0.5 1.5 2.5 3.5 4.5 ... 356.5 357.5 358.5 359.5
* y (y) float32 -78.39 -78.19 -77.98 -77.77 ... 88.96 89.37 89.74
lat (y, x) float32 dask.array<chunksize=(291, 360), meta=np.ndarray>
* lev (lev) float64 3.047 9.454 16.36 ... 5.375e+03 5.625e+03
lev_bounds (lev, bnds) float64 dask.array<chunksize=(45, 2), meta=np.ndarray>
lon (y, x) float32 dask.array<chunksize=(291, 360), meta=np.ndarray>
* time (time) int64 0 708 1416 2148 ... 1443192 1443924 1444656
time_bounds (time, bnds) float64 dask.array<chunksize=(1980, 2), meta=np.ndarray>
lat_verticies (y, x, vertex) float32 dask.array<chunksize=(291, 360, 4), meta=np.ndarray>
lon_verticies (y, x, vertex) float32 dask.array<chunksize=(291, 360, 4), meta=np.ndarray>
* bnds (bnds) int64 0 1
* vertex (vertex) int64 0 1 2 3
lon_bounds (bnds, y, x) float32 dask.array<chunksize=(1, 291, 360), meta=np.ndarray>
lat_bounds (bnds, y, x) float32 dask.array<chunksize=(1, 291, 360), meta=np.ndarray>
* member_id (member_id) <U8 'r1i1p1f1'
Data variables:
thetao (member_id, time, lev, y, x) float32 dask.array<chunksize=(1, 6, 45, 291, 360), meta=np.ndarray>
Attributes:
CCCma_model_hash: 3dedf95315d603326fde4f5340dc0519d80d10c0
CCCma_parent_runid: rc3-pictrl
CCCma_pycmor_hash: 094aa868e787693cfe55b2f1665f6a6b0880b03a
CCCma_runid: rc3.1-his01
Conventions: CF-1.7 CMIP-6.2
YMDH_branch_time_in_child: 1850:01:01:00
YMDH_branch_time_in_parent: 5201:01:01:00
activity_id: CMIP
branch_method: Spin-up documentation
branch_time_in_child: 0.0
branch_time_in_parent: 1223115.0
cmor_version: 3.4.0
contact: ec.cccma.info-info.ccmac.ec@canada.ca
creation_date: 2019-03-14T05:00:40Z
data_specs_version: 01.00.29
experiment: all-forcing simulation of the recent past
experiment_id: historical
external_variables: areacello volcello
forcing_index: 1
frequency: mon
further_info_url: https://furtherinfo.es-doc.org/CMIP6.CCCma.C...
grid: ORCA1 tripolar grid, 1 deg with refinement t...
grid_label: gn
history: 2019-03-14T05:00:40Z ;rewrote data to be con...
initialization_index: 1
institution: Canadian Centre for Climate Modelling and An...
institution_id: CCCma
license: CMIP6 model data produced by The Government ...
mip_era: CMIP6
nominal_resolution: 100 km
parent_activity_id: CMIP
parent_experiment_id: piControl
parent_mip_era: CMIP6
parent_source_id: CanESM5
parent_time_units: days since 1850-01-01 0:0:0.0
parent_variant_label: r1i1p1f1
physics_index: 1
product: model-output
realization_index: 1
realm: ocean
references: Geophysical Model Development Special issue ...
source: CanESM5 (2017): \naerosol: interactive\natmo...
source_id: CanESM5
source_type: AOGCM
sub_experiment: none
sub_experiment_id: none
table_id: Omon
table_info: Creation Date:(13 December 2018) MD5:e84cb97...
title: CanESM5 output prepared for CMIP6
tracking_id: hdl:21.14100/5446945e-4a3c-43a1-babd-af08607...
variable_id: thetao
variant_label: r1i1p1f1
version: v20190306
status: 2019-11-06;created;by nhn2@columbia.edu
netcdf_tracking_ids: hdl:21.14100/5446945e-4a3c-43a1-babd-af08607...
version_id: v20190306
intake_esm_varname: ['thetao']
intake_esm_dataset_key: CMIP.CCCma.CanESM5.historical.Omon.gn
The vertex convention is consistent across models. The points are sorted from lower-left, upper-left, upper-right to lower-right.
## TL;DR How to put it all together
To combine all these (or just some you like), you can create a wrapper function as above, or you can use the provided combined_preprocessing, which does all the above. > Due to concerns regarding the dask performance of large datasets, the latest version of combined_preprocessing does not apply replace_x_y_nominal_lat_lon anymore. You can still apply this at a later point, preferrably at the end of a processing step, to enable rough
selection of regions (see example below).
[23]:
from xmip.preprocessing import combined_preprocessing
# lets load a bunch more models this time
query = dict(experiment_id=['piControl', 'historical'],
table_id='Oyr',
source_id=[
'GFDL-ESM4',
'IPSL-CM6A-LR',
'CanESM5',
'CanESM5-CanOE',
'MPI-ESM-1-2-HAM',
'MPI-ESM1-2-HR',
'MPI-ESM1-2-LR',
'ACCESS-ESM1-5',
'MRI-ESM2-0',
'IPSL-CM5A2-INCA',
'EC-Earth3-CC'
],
variable_id='o2',
grid_label=['gn', 'gr'])
cat = col.search(**query)
print(cat.df['source_id'].unique())
with dask.config.set(**{'array.slicing.split_large_chunks': True}):
dset_dict = cat.to_dataset_dict(zarr_kwargs=z_kwargs,
preprocess=combined_preprocessing)
['GFDL-ESM4' 'IPSL-CM6A-LR' 'CanESM5' 'CanESM5-CanOE' 'MPI-ESM-1-2-HAM'
'MPI-ESM1-2-HR' 'MPI-ESM1-2-LR' 'ACCESS-ESM1-5' 'MRI-ESM2-0'
'IPSL-CM5A2-INCA' 'EC-Earth3-CC']
--> The keys in the returned dictionary of datasets are constructed as follows:
'activity_id.institution_id.source_id.experiment_id.table_id.grid_label'
[24]:
fig, axarr = plt.subplots(nrows=4, ncols=3, figsize=[25,15])
for ax,(k, ds) in zip(axarr.flat,dset_dict.items()):
if 'member_id' in ds.dims:
ds = ds.isel(member_id=0)
da = ds.o2.isel(time=0).interp(lev=2500)
# this step is necessary to order the longitudes properly for simple plotting. Alternatively you could use a proper map projection
# with e.g. cartopy and would not need this step
da = replace_x_y_nominal_lat_lon(da)
da = da.sel(x=slice(100, 200), y=slice(-20,20))
try:
da.plot(ax=ax, vmax=0.25, vmin=0.05)
except:
print(k)
pass
ax.set_title(k)
Appeal¶
``xmip`` is still under very active development, and its true strength will be to ‘crowdsource’ fixes for common problems. If you notice errors, please try to isolate them by applying the single untility functions and then raise an issue on github
