Masking out ocean basins¶
xmip is able to use and aggregate regionmask provided by Natural Earth via the regionmask package.
This functionality needs the upstream master of regionmask: > In a cell do !pip install git+https://github.com/mathause/regionmask.git and restart the notebook
Also, xmip is currently not installed by default: > In a cell do !pip install git+https://github.com/jbusecke/xmip.git and restart the notebook
[1]:
from dask.distributed import Client
client = Client("tcp://10.32.0.154:42411")
client
[1]:
Client
|
Cluster
|
[1]:
# !pip install git+https://github.com/mathause/regionmask.git
# !pip install -e ../
[2]:
import regionmask
import intake
import matplotlib.pyplot as plt
from xmip.preprocessing import combined_preprocessing
import xarray as xr
import numpy as np
/srv/conda/envs/notebook/lib/python3.7/site-packages/intake/source/discovery.py:136: FutureWarning: The drivers ['geojson', 'postgis', 'shapefile', 'spatialite'] do not specify entry_points and were only discovered via a package scan. This may break in a future release of intake. The packages should be updated.
FutureWarning)
Lets import ocean temperature data from a variety of models to see if the masking works well.
[3]:
# import example cloud datasets
col_url = "https://raw.githubusercontent.com/NCAR/intake-esm-datastore/master/catalogs/pangeo-cmip6.json"
col = intake.open_esm_datastore(col_url)
cat = col.search(source_id=['CAMS-CSM1-0', 'CNRM-CM6-1', 'CNRM-ESM2-1', 'ACCESS-CM2', 'ACCESS-ESM1-5', 'EC-Earth3-Veg',
'MIROC-ES2L', 'MIROC6', 'HadGEM3-GC31-LL', 'UKESM1-0-LL', 'MPI-ESM1-2-HR', 'MRI-ESM2-0',
'NorCPM1', 'GFDL-CM4', 'GFDL-ESM4', 'NESM3'],
experiment_id='historical', variable_id='thetao')
data_dict = cat.to_dataset_dict(zarr_kwargs={'consolidated': True, 'decode_times': False},
preprocess=combined_preprocessing)
--> The keys in the returned dictionary of datasets are constructed as follows:
'activity_id.institution_id.source_id.experiment_id.table_id.grid_label'
--> There is/are 20 group(s)
[######## ] | 20% Completed | 2.7sMIROC6: No units found
MIROC-ES2L: No units found
[#################### ] | 50% Completed | 3.8sMIROC-ES2L: No units found
MIROC6: No units found
[#################### ] | 50% Completed | 4.5sMIROC6: No units found
[#################### ] | 50% Completed | 4.7sMIROC-ES2L: No units found
[######################## ] | 60% Completed | 5.3sMIROC6: No units found
[######################## ] | 60% Completed | 5.6sMIROC6: No units found
[########################## ] | 65% Completed | 6.6sMIROC6: No units found
[############################ ] | 70% Completed | 7.4sMIROC6: No units found
[############################## ] | 75% Completed | 7.8sMIROC6: No units found
[############################## ] | 75% Completed | 8.1sMIROC6: No units found
[################################## ] | 85% Completed | 8.6sMIROC6: No units found
[########################################] | 100% Completed | 11.1s
First we need to load the natural earth ocean basins.
[4]:
# load ocean basin data
basins = regionmask.defined_regions.natural_earth.ocean_basins_50
basins.plot(add_ocean=False, add_label=False)
[4]:
<cartopy.mpl.geoaxes.GeoAxesSubplot at 0x7f84c86d0cd0>
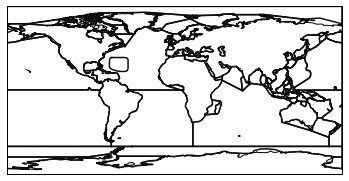
This is a neat dataset, but for large scale climate analysis it is often useful to separate the large scale ocean basins (e.g. the full Pacific, with all marginal seas included). For this we can use merge_mask.
[5]:
from xmip.regionmask import merged_mask
When using the defaults this will index the North and South Pacific with numeric values of 2 and 3 respectively. The Maritime Continent gets a separate index (4). We will now load example data from many CMIP6 models, create the mask, and select only the points with values 2,3, and 4.
[6]:
# loop over models and mask out the pacific
import cartopy.crs as ccrs
for k, ds in data_dict.items():
if 'lev' in ds.dims:
model = ds.attrs['source_id']
if 'member_id' in ds.dims:
ds = ds.isel(member_id=0)
ds = ds.thetao.isel(time=0, lev=0).squeeze()
mask = merged_mask(basins,ds)
kwargs = dict(x='lon', y='lat',transform = ccrs.PlateCarree(), infer_intervals=False)
fig, (ax1, ax2, ax3) = plt.subplots(ncols=3, figsize=[20,2], subplot_kw={'projection':ccrs.Robinson(190)})
ds.plot(ax=ax1, **kwargs)
ax1.set_title(f"Raw field {model}")
ds_masked = ds.where(np.logical_or(np.logical_or(mask == 2, mask==3),mask==4)) # Pacific + Maritime Continent
ds_masked.plot(ax=ax2, **kwargs)
ax2.set_title(f"Masked Pacific {model}")
mask.plot(ax=ax3, cmap='tab20', vmin=0, vmax=19, **kwargs)
ax3.set_title(f"Full Mask {model}")
for ax in [ax1, ax2, ax3]:
ax.coastlines()
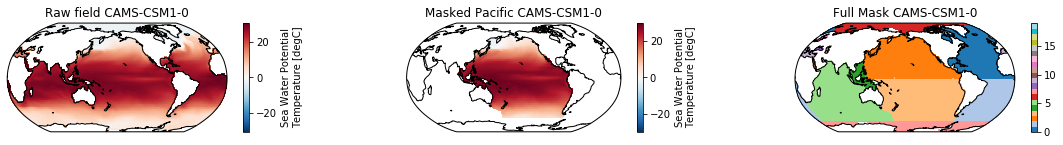
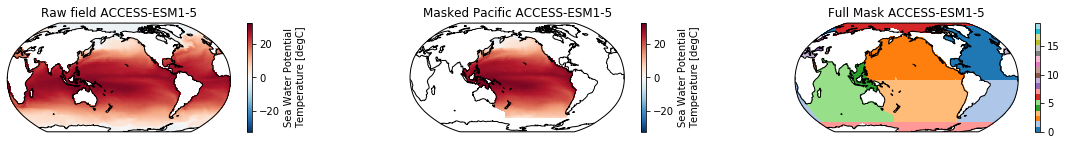
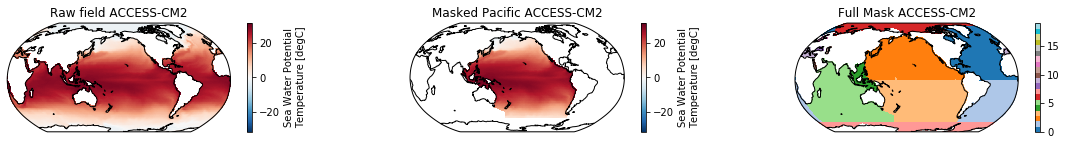
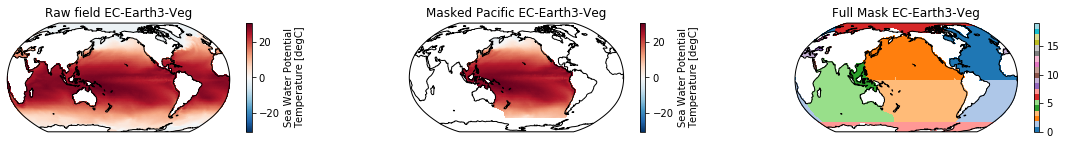
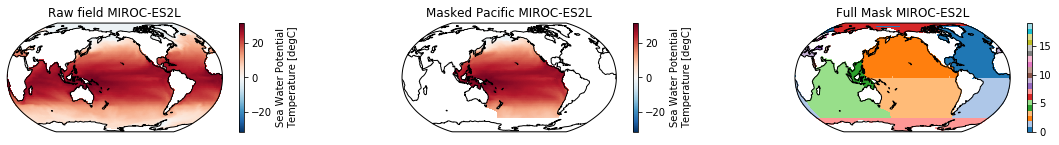
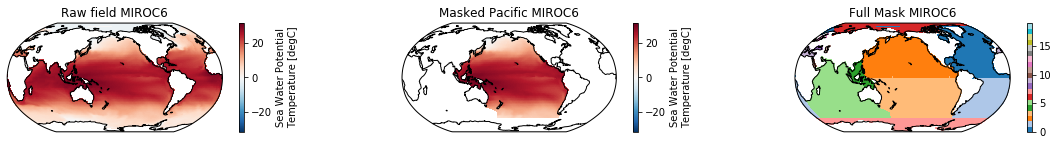
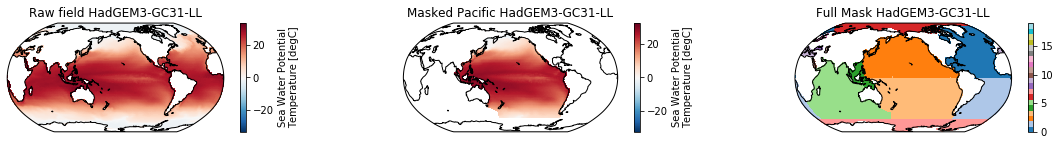
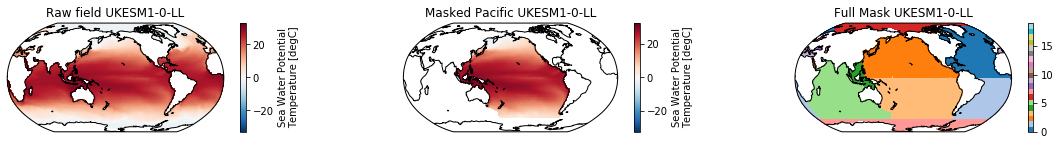
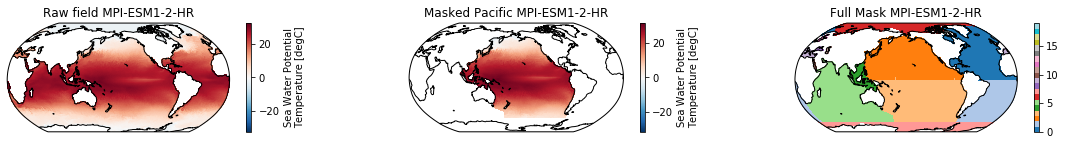
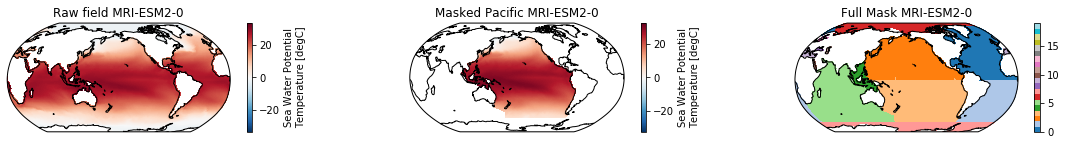
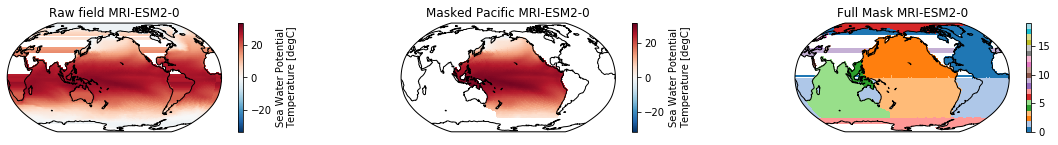
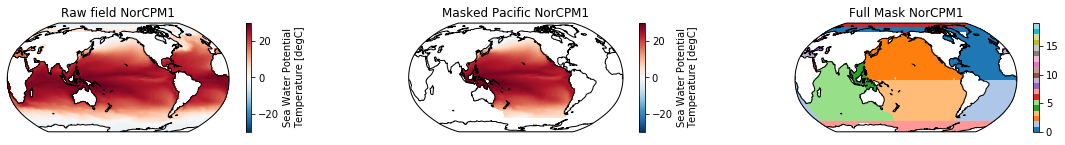
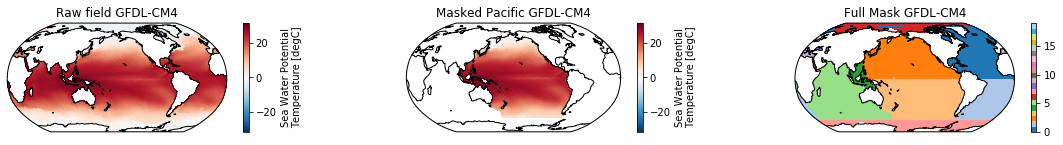
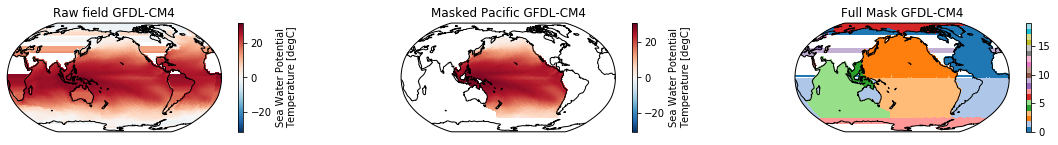
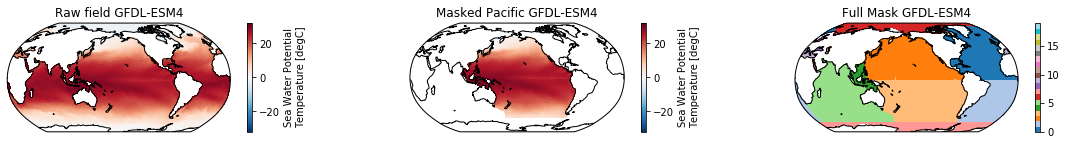
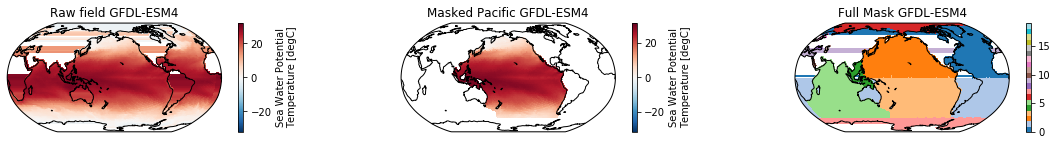
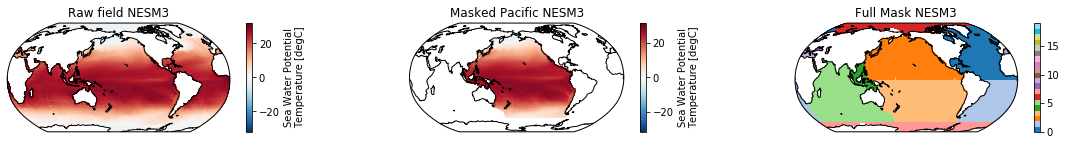
The masked dataset now only contains values in the Pacific and can be further manipulated.
[ ]:
